FIGURE 6.
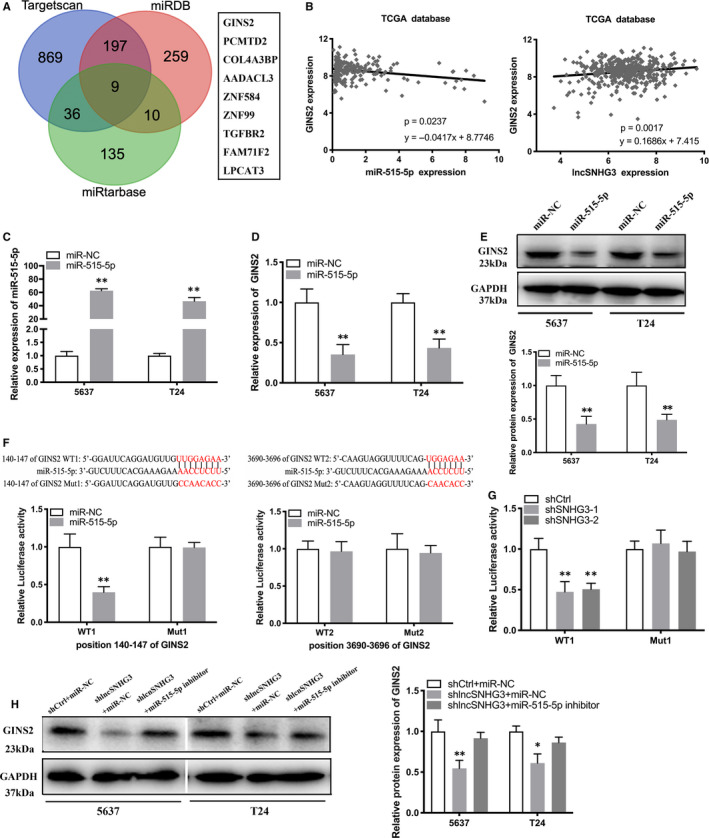
lncRNA SNHG3 positively modulated GINS2 expression via sponging miR‐515‐5p. A, Original data obtained from bioinformatics databases predicted 9 candidate genes targeted by miR‐515‐5p. B, Statistical correlation analysis showed that miR‐515‐5p expression was negatively correlated with GINS2 expression in Bca tissues, whereas lncRNA SNHG3 expression was positively correlated with GINS2 expression in Bca tissues. C, Efficiency of miR‐515‐5p mimic was confirmed by qRT‐PCR. (D and E) qRT‐PCR and Western blotting assay demonstrated that miR‐515‐5p expression levels were negatively correlated with GINS2 expression levels in Bca cells. F, Bioinformatics analysis predicted two probable 3’UTR sequences of GINS2 (WT1 and WT2) complemented to the seed sequence of miR‐515‐5p. Dual‐luciferase reporter assay showed that co‐transfection of WT1 and miR‐515‐5p significantly inhibited luciferase activity. G, Dual‐luciferase reporter assay determined that co‐transfection of WT1 and shlncRNA SNHG3 significantly inhibited luciferase activity. H, Reduced miR‐515‐5p reversed GINS2 expression inhibition induced by shlncRNA SNHG3 in Bca cells. Data are represented as mean ± SD. *P < 0.05; **P < 0.01
