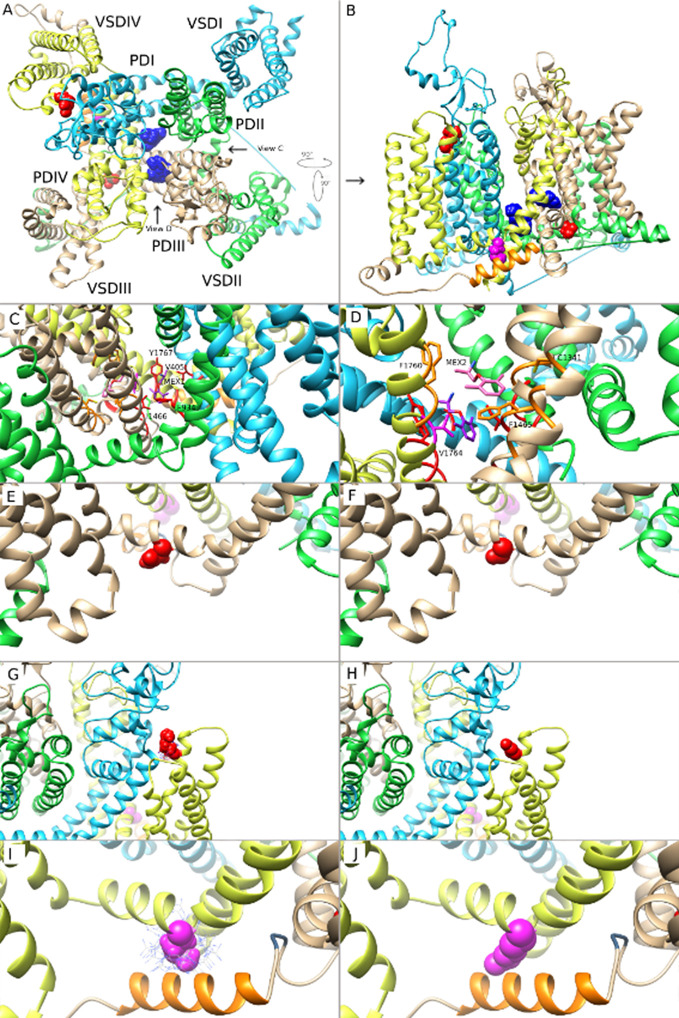
Figure 7.
Molecular modeling of mexiletine (MEX) binding to MEX-sensitive (N1325S, R1623Q) and -insensitive (M1652R) mutant channels. (A) A molecular model of wild-type (WT) Nav1.5 in inactivated state, viewed from the extracellular side of the channel with MEX bound at two possible positions. (B) Side view of the channel with the three amino acids of interest facing the reader for clarity. (C) Closer view of MEX binding site 1. A single amino acid side chain in each binding region (S6 segments from all 4 domains) are labeled and colored. (D) Closer view of MEX binding site 2. Labels are specific to MEX binding site 2. A single amino acid side chain in each binding region (DIII-S5, DIII-S6, and DIV-S6) is labeled and shown. (E, G, I) Closer view of the three amino acid residues N1325, R1623, and M1652, respectively. (F, H, J). Closer view of the optimal orientation for N1325S, R1623Q, and M1652R substitutions, respectively. Blue dots in panels (I, J) represent possible orientations of the M1652R substitution.