Figure 6. Nucleosome Remodeling and Bypass during the Homology Search.
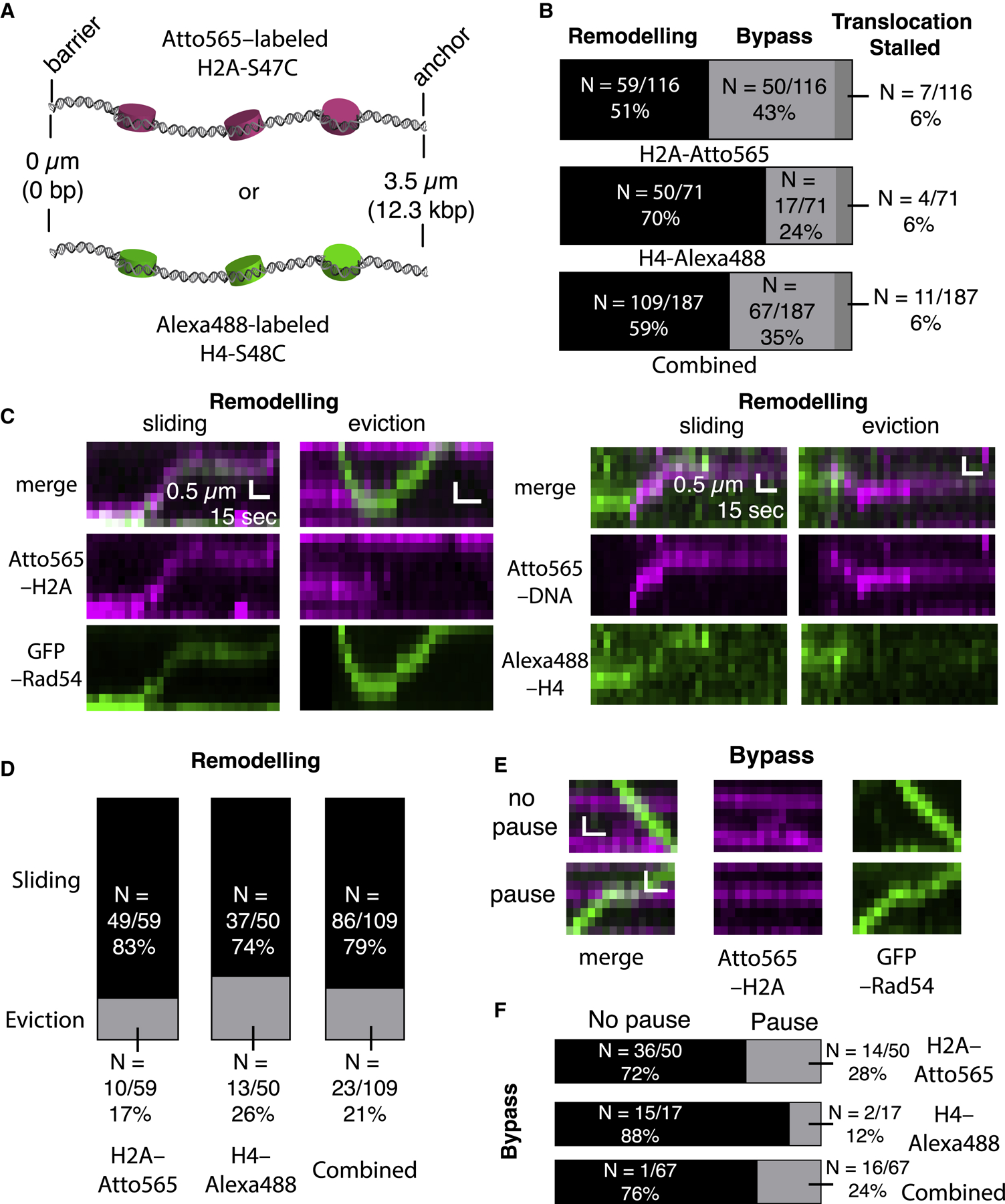
(A) Schematic of nucleosome-bound donor dsDNA substrates labeled with Atto565-H2A or Alexa 488-H4.
(B) Summary of different outcomes during PSC encounters with single nucleosomes for labeled H2A or labeled H4 with PSCs prepared with the tailed duplex (21-nt ssDNA) substrate. The bottom graph shows the combined labeled H2A and labeled H4 datasets.
(C) Kymographs showing examples Atto565-H2A nucleosomes (magenta) being remodeled by PSCs labeled with GFP-Rad54 (green, left panels) and examples of Alexa 488-H4 nucleosomes (green) being remodeled by PSCs labeled with Atto565-labeled tailed duplex DNA (magenta, right panels).
(D) Distributions of nucleosome remodeling events, sliding or eviction, for H2A-Atto565-labeled, H4-Alexa 488-labeled, and combined datasets.
(E) Kymographs depicting nucleosomes (magenta) being bypassed by translocating PSCs (green) with or without evident PSC pausing at the nucleosome.
(F) Fraction of bypass events where the PSCs pause during nucleosome bypass for nucleosomes labeled with H2A-Atto565, H4-Alexa 488, and combined datasets.
