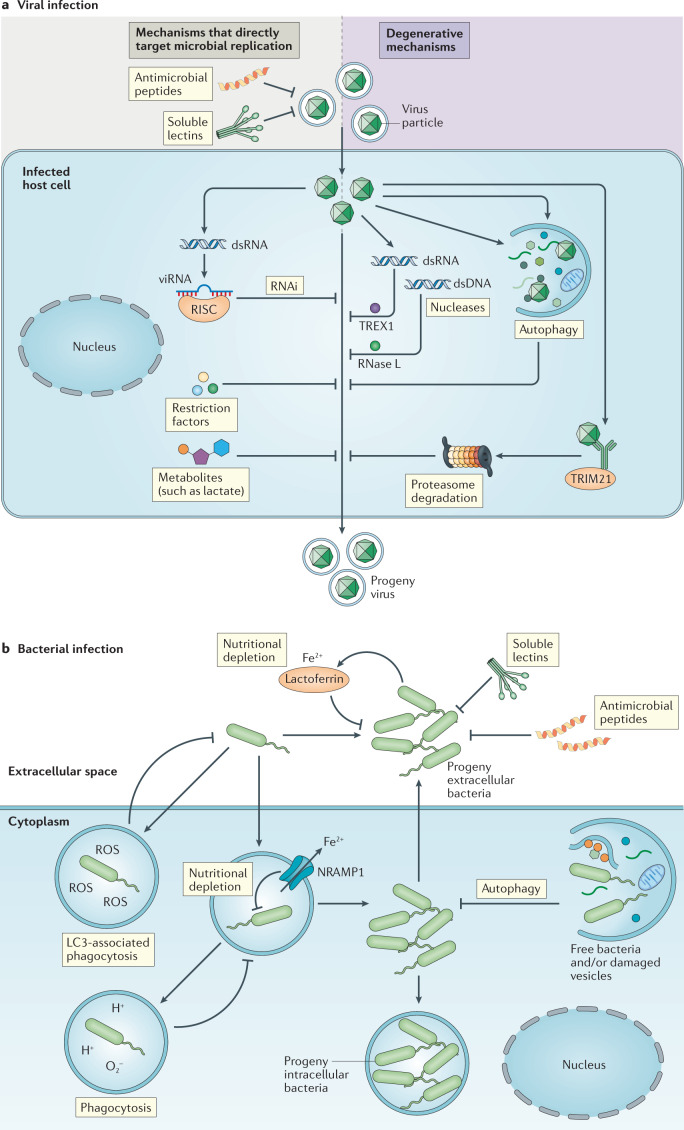
Fig. 3. Overview of the regulation of microbial replication by constitutive innate immune mechanisms.
a | Constitutive innate immune mechanisms and viral infection. The accumulation of specific viral molecular structures (such as double-stranded RNA (dsRNA) or capsids) and cellular stress responses (such as autophagy) activate constitutive–latent mechanisms with direct antiviral activity, independently of pattern recognition receptors. Some of the antiviral effector functions target microbial replication by blocking specific steps in the replication cycles of viruses; these effectors include soluble lectins, antimicrobial peptides, restriction factors, RNA interference (RNAi) and metabolites. Other antiviral effectors of the constitutive response function through the degradation of virus particles; these include nucleases such as TREX1, which degrades viral DNA in the cytoplasm, and RNase L, which degrades viral RNA, as well as autophagy and proteasomal degradation. Viruses can be targeted for proteasomal degradation by the ubiquitin E3 ligase TRIM21, which binds to antibody-attached viral capsids. b | Constitutive innate immune mechanisms and bacterial infection. The presence of bacteria changes the local microenvironment, for example through the accumulation of hydrophobic and charged bacterial surfaces or alteration of cellular metabolism. This activates antibacterial activities independently of pattern recognition receptors, including inactivation by soluble lectins and antimicrobial peptides, nutritional depletion by natural resistance-associated macrophage protein 1 (NRAMP1) and lactoferrin, and bacterial degradation by phagocytosis and basal autophagy. dsDNA, double-stranded DNA; RISC, RNA-induced silencing complex; ROS, reactive oxygen species; viRNA, virus-derived small interfering RNA.