Figure 2.
Deep Mutational Scanning of All Amino Acid Mutations to the SARS-CoV-2 RBD
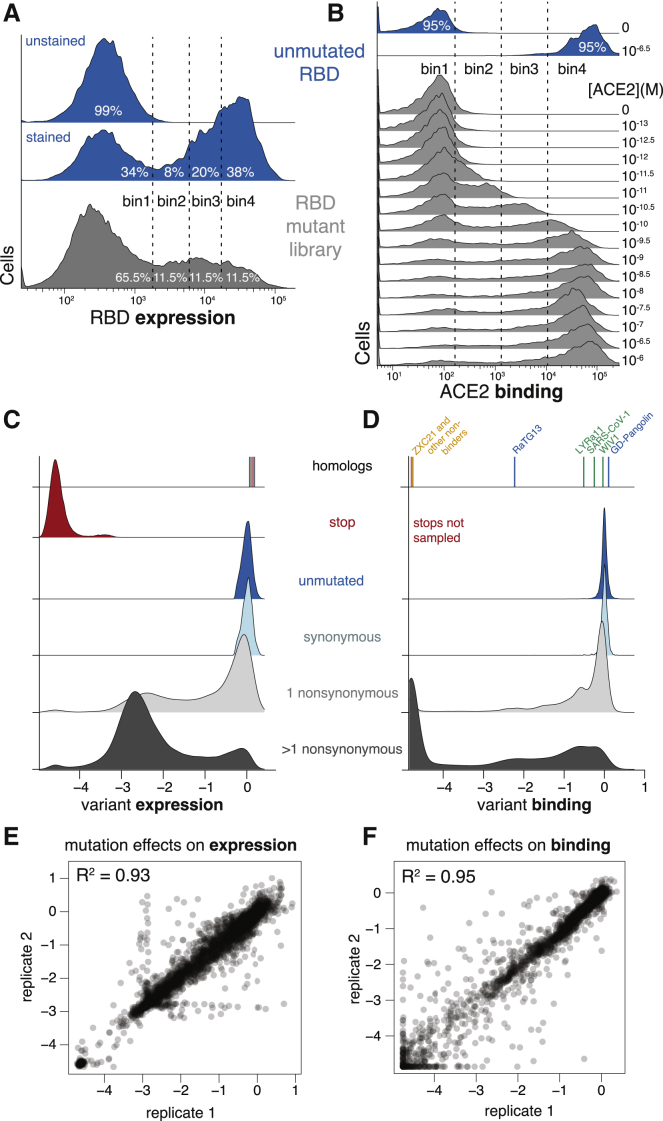
(A and B) FACS approach for deep mutational scans for expression (A) and binding (B). Cells were sorted into four bins from low to high expression or binding, with separate sorts for each ACE2 concentration. The frequency of each library variant in each bin was determined by Illumina sequencing of the barcodes of cells collected in that bin, enabling reconstruction of per-variant expression and binding phenotypes. Bin boundaries were drawn based on distributions of expression or binding for unmutated SARS-CoV-2 controls (blue), and gray shows the distribution of library variants for library replicate 1 in these bins.
(C and D) Distribution of library variant phenotypes for expression (C) and binding (D), with variants classified by the types of mutations they contain. Internal control RBD homologs are indicated with vertical lines, colored by clade as in Figure 1A. Stop-codon-containing variants were purged by an RBD+ pre-sort prior to ACE2 binding measurements and so are not sampled in (D).
(E and F) Correlation in single-mutant effects on expression (E) and binding (F), as determined from independent mutant library replicates.
See also Figures S1 and S2 and Table S1.