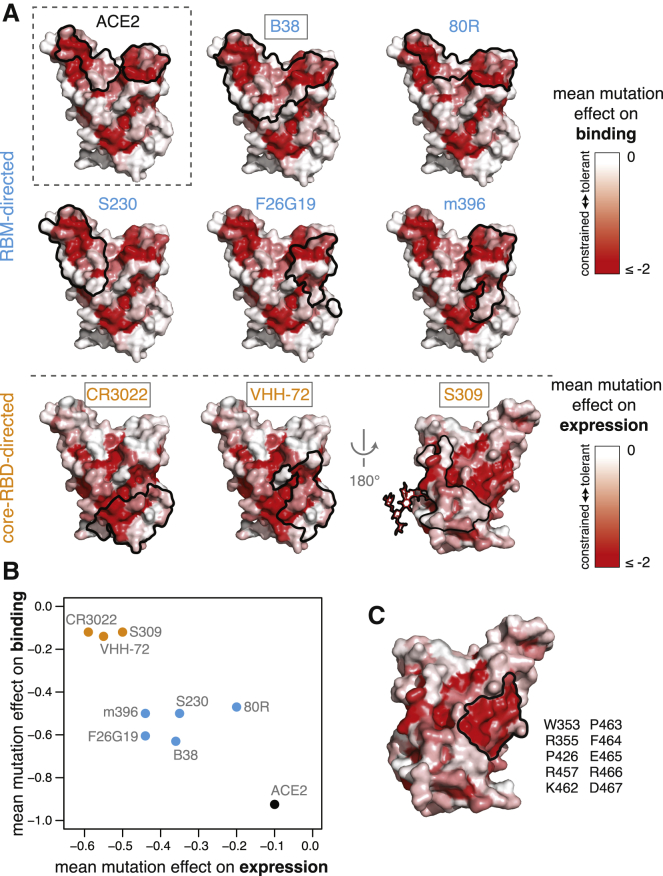
Figure 6.
Mutational Constraint of Antibody Epitopes
(A) For ACE2 and each of 8 RBD-directed antibodies, black outlines indicate the epitope structural footprint, with surfaces colored by mutational constraint (red indicates more constrained). Names of antibodies capable of neutralizing SARS-CoV-2 are boxed. Constraint is illustrated as mutational effects on binding for RBM-directed antibodies (blue, top) and expression for core-RBD-directed antibodies (orange, bottom). The N343 glycan, which is present in the S309 epitope and is constrained with respect to expression, is shown only on this surface for clarity.
(B) Average mutational constraint for binding and expression within each epitope. Points are colored according to the RBM versus core-RBD designation in (A).
(C) Identification of a patch of mutational constraint surrounding RBD residue E465, which has not yet been targeted by any described antibodies. Surface is colored according to mutational effects on expression, as in (A, bottom). Residues in this constrained E465 patch are listed. See also Figure S6.