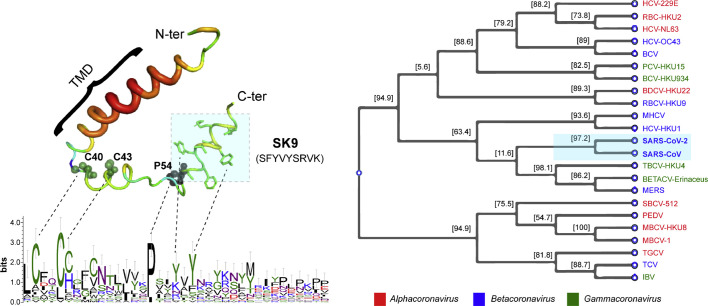
Fig. 5.
The conserved sequence analyses of the SARS-CoV-2 E protein with other members of the family. The left panel shows a multiple sequence alignment of several different CoV E proteins. The highly conserved Cysteine and Proline residues are labeled and represented with a sphere. The amyloidogenic SK9 fragment and TMD are highlighted in the figure. The multiple sequence alignment of proteins were carried out using ClustalW2 at the European Bioinformatics Institutes server, and WebLogo 3 was used to generate the figure. The right panel represents a Phylogenetic tree, based on amino acid sequences for E proteins of the CoV family. The tree was built using Mega X software based on maximum likelihood. Multiple sequence alignment was performed using the clustal method. The following full length amino acid sequences were used: Human coronavirus 229E (HCV-229E), Human coronavirus NL63 (HCV-NL63), Miniopterus bat coronavirus 1 (MBCV-1), Miniopterus bat coronavirus HKU8 (MBCV-HKU8), Porcine epidemic diarrhea virus (PEDV), Rhinolophus bat coronavirus HKU2 (RBC-HKU2), Scotophilus bat coronavirus 512 (SBCV-512), Murine hepatitis virus strain JHM (MHCV), Bovine coronavirus (BCV), Human coronavirus OC43 (HCV-OC43), Betacoronavirus Erinaceus/VMC/DEU/2012 (BetaCV-Erinaceus), Human coronavirus HKU1 (HCV-HKU1), Middle East respiratory syndrome-related coronavirus (MERS), Rousettus bat coronavirus HKU9 (RBCV-HKU9), Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), Severe acute respiratory syndrome-related coronavirus (SARS-CoV), Tylonycteris bat CoV HKU4 (TBCV-HKU4), Infectious bronchitis virus (IBV), Turkey coronavirus (TCV), Bottlenose dolphin coronavirus HKU22 (BDCV-HKU22), Bulbul coronavirus HKU11–934 (BCV-HKU-934), Porcine coronavirus HKU15 (PCV-HKU15). Genus of the viruses is colour coded in the figure.