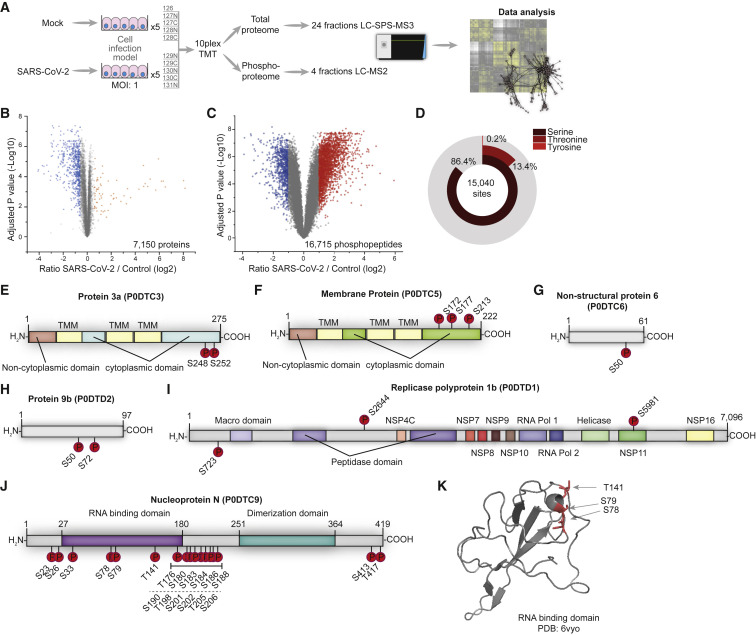
Figure 1.
Phosphoproteomic Profiling of SARS-CoV-2-Infected Cells
(A) Experimental scheme. Caco-2 cells were infected with SARS-CoV-2 for 1 h (MOI 1), washed, and incubated for an additional 24 h. Proteins were extracted and prepared for bottom-up proteomics. All 10 conditions were multiplexed using TMT10 reagents. A total of 250 μg pooled samples were used for whole-cell proteomics (24 fractions) and the remainder (~1 mg) enriched for phosphopeptides by Fe-NTA. Phosphopeptides were fractionated into 8 fractions and concatenated into 4 fractions. All of the samples were measured on an Orbitrap Fusion Lumos.
(B) Volcano plot showing fold changes (FCs) of infected versus mock cells for all quantified phosphopeptides. p values were calculated using an unpaired, 2-sided Student’s t test with equal variance assumed and adjusted using the Benjamini-Hochberg false discovery rate (FDR) method (N = 5 biological replicates). Orange or blue points indicate significantly increased or decreased phosphopeptides, respectively.
(C) Volcano plot showing differences between SARS-CoV-2 and mock-infected cells in total protein levels for all quantified proteins. p values were calculated using an unpaired, 2-sided Student’s t test with equal variance assumed and adjusted using the Benjamini-Hochberg FDR method (N = 5). Orange or blue points indicate significantly increased or decreased phosphopeptides, respectively.
(D) Distribution of phosphorylation sites identified across modified amino acids. See also Figure S1 and Tables S1 and S2.
(E–K) Domain structures of SARS-CoV-2 proteins predicted by InterPro. Identified phosphorylation sites are indicated. Protein 3a (E), membrane protein M (F), non-structural protein 6 (G), protein 9b (H), replicase polyprotein 1b (I), and nucleoprotein N (J). (K) X-ray structure of the RNA-binding domain (PDB: 6vyo, residues 47–173), with identified phosphorylation sites marked in red.