Figure 3.
Drug-Target Phosphoprotein Network Analysis Identifies Growth Factor Signaling as Central Hub for Possible Intervention by Repurposed Drugs
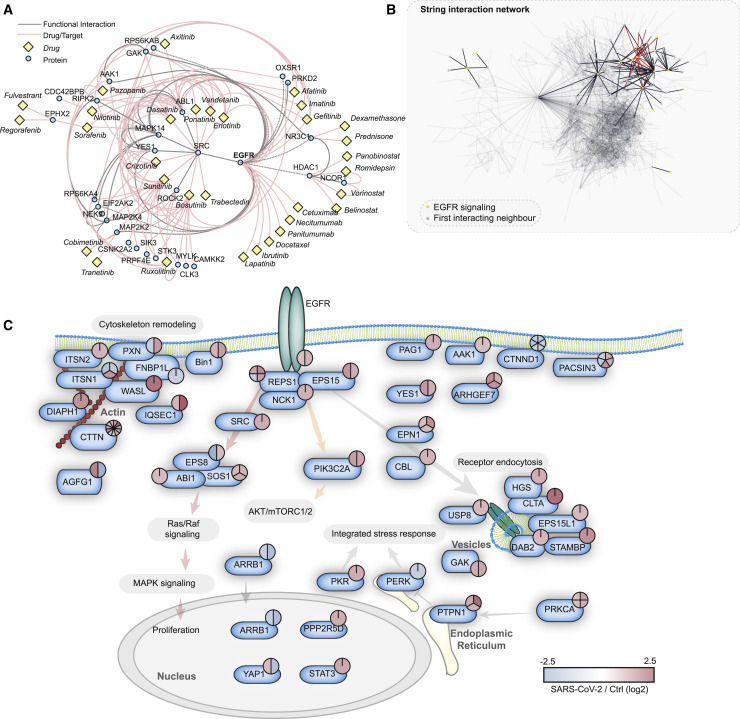
(A) Proteins significantly increased in phosphorylation (FC > 1, FDR < 0.05) were subjected to ReactomeFI pathway analysis and overlaid with a network of US Food and Drug Administration (FDA)-approved drugs. The network was filtered for drugs and drug targets only, to identify pathways that could be modulated by drug repurposing. The red lines indicate drug-target interactions, and the gray lines protein-protein interactions. The identified drugs are represented with yellow rectangles, while proteins are represented by blue circles.
(B) Search across all proteins with significant phosphorylation changes upon SARS-CoV-2 infection for proteins related to the EGFR pathway. The STRING network highlights all of the proteins annotated for EGFR signaling and their direct interaction neighbors. The red lines indicate direct EGFR interactions, the black lines indicate interactions between pathway members, and the gray lines represent filtered interactions to represent the whole network.
(C) Pathway representation of proteins identified in (B) to be direct functional interactors of EGFR, according to the STRING interaction database (confidence cutoff 0.9). The phosphorylation changes of all significantly regulated sites are indicated by color-coded pie charts. Red indicates upregulation and blue indicates downregulation.
See also Figure S4.