Fig. 1 |. Using balancer chromosomes to probe the relationship between genome architecture and function.
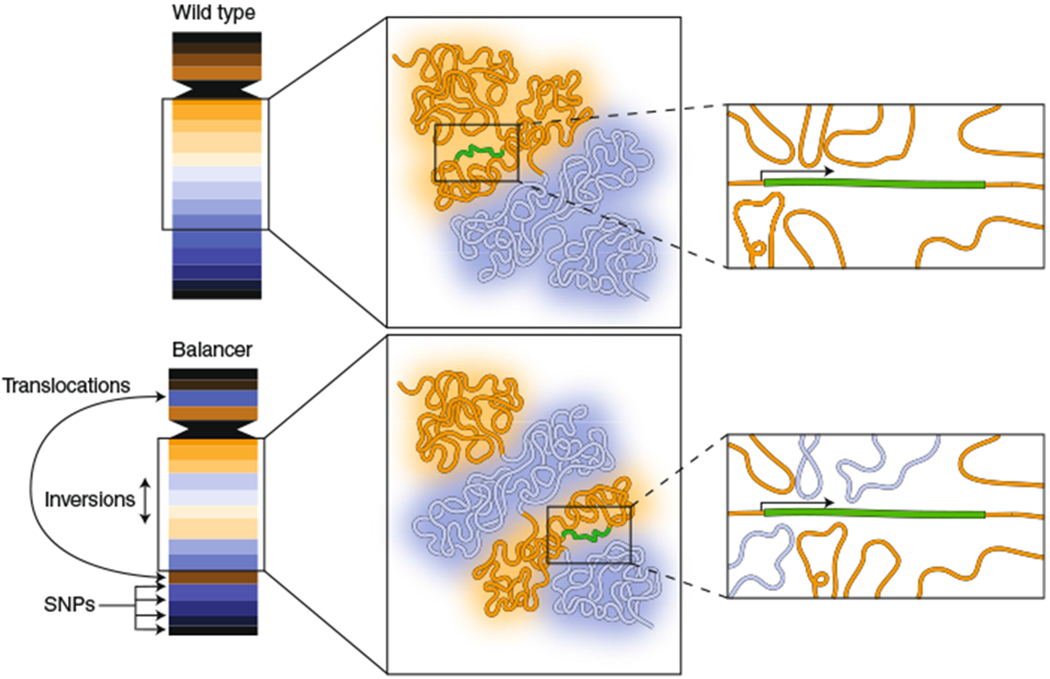
Balancer chromosomes contain inversions, deletions and translocations, thus making them topologically distinct from wild-type chromosomes. They also contain single-nucleotide variants, enabling capture of allele-specific expression data from RNA-seq. Ghavi-Helm et al.1 took advantage of the presence of wild-type and balancer chromosomes in the same nucleus to compare the effects of altered genome topology on gene expression. The activity of many genes was found to be insensitive to the chromatin topology in their environment. As an example, an internal inversion of two chromatin domains (blue, orange) may expose a gene (green) to distinct chromatin interactions on the balancer chromosome, yet its activity is unaffected. The results may suggest that multiple enhancers, often located in a single chromatin domain, act coordinately on a single promoter and that the effect of a given enhancer can be compensated for by other enhancers, even though it may originate from a distinct chromatin domain.
