Figure 2.
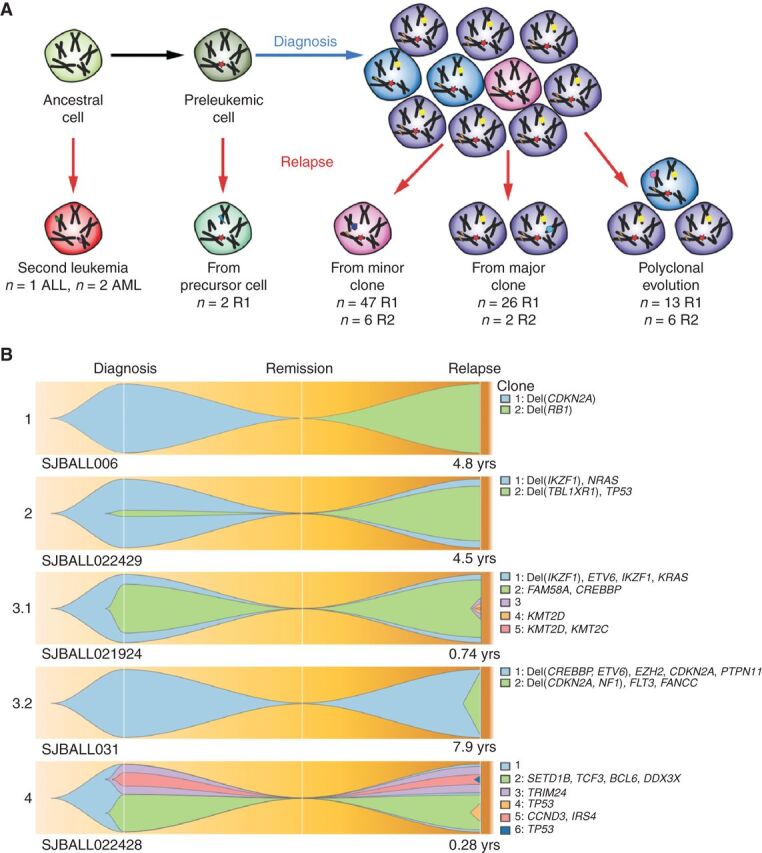
Patterns of relapse in ALL. A, Schematic overview of mechanisms of clonal evolution. Three patients developed a second primary tumor that was not clonally related to the previous tumor occurrence. Two patients developed a tumor that shared only one founding fusion between diagnosis and relapse, indicating the disease relapsed from a preleukemic cell. Further relapses arose through evolution from a minor clone, a major clone, or multiple clones. B, Fish plots of the clonal evolution models inferred from somatic mutations detected in diagnosis and relapse samples. MAF of the somatic mutations was used by the sciClone R package (14) to infer potential clonal clusters (shown in different colors) and visualized by Fishplot (ref. 13; see Methods). Four major clonal evolution models were observed: 1, relapse sample is a second primary leukemia with no somatic mutations shared with diagnosis; 2, a minor clone (somatic mutations' median MAF of the clone is less than 30%) in diagnosis develops into the major clone in relapse; 3, a major clone (somatic mutations' median MAF of the clone is greater than 30%) is preserved from diagnosis to relapse (3.1) or emerges as a major clone at relapse (3.2); 4, multiple subclones in diagnosis develop as multiple subclones in relapse. For each exemplary case, the time from diagnosis to relapse is indicated. Focal deletions and nonsilent somatic mutations on cancer genes (according to the COSMIC Cancer Gene Census and well-known leukemia relevant genes) for each inferred clone are shown on the right side.
