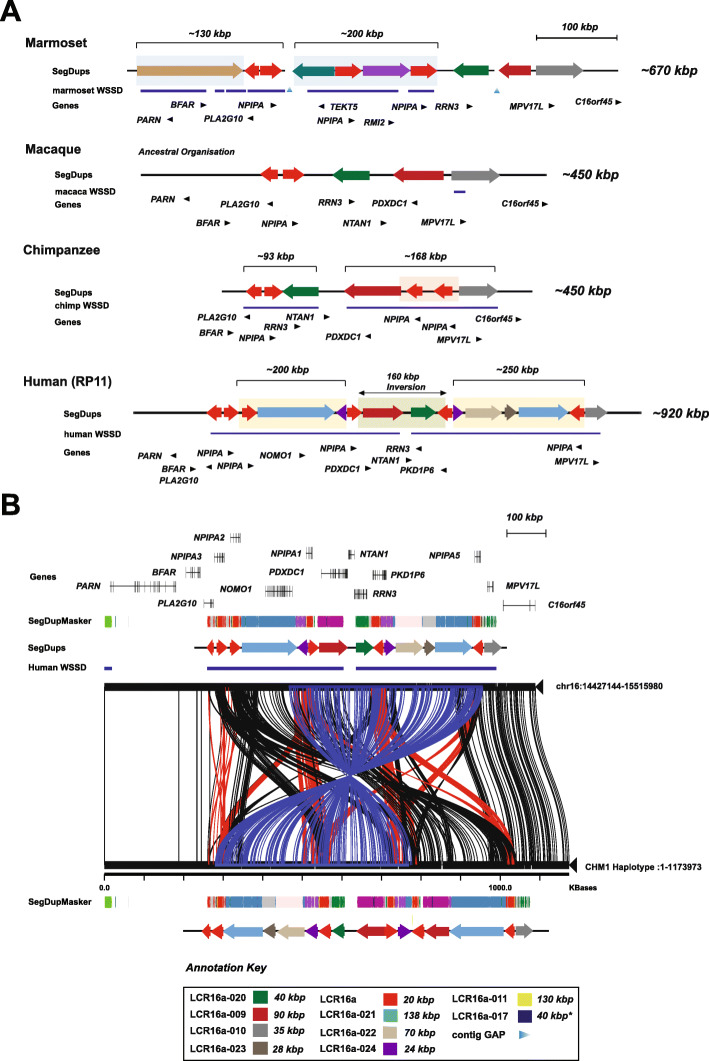
Fig. 4.
Structure of the ancestral chromosome 16p13 locus. The structure and organization of chr16p13 in four primate lineages is shown based on sequencing of a tiling path of BAC clones for each primate haplotype. SDs (colored arrows) and gene models (black arrows) are shown with respect to lineage-specific duplications (blue bars) identified based on sequence read-depth (WSSD) [1]. a The chromosome 16p13 region has expanded and contracted hundreds of kilobases due to lineage-specific duplication. Note the ancestral ~ 160-kbp inversion between the human RP11 haplotype and all other primates. The ancestral LCR16a duplicon in macaque shows a single copy of NPIP, compared to three copies in marmoset and chimpanzee, and five copies in human. b A Miropeats comparison between two human haplotypes at the ancestral locus on chr16p13. CHM1 BACs tiling across the chr16p13 region were sequenced and assembled using PacBio SMRT sequencing to create a super contig. The SD organization is depicted using colored arrows. Miropeats between RP11 and CHM1 contigs shows pairwise differences between orthologous regions. A ~ 400-kbp inversion is detected in the CHM1 haplotype, flanked by LCR16a core duplicons (blue lines). CHM1 also carries an additional duplication corresponding to LCR16a-009, which contains PDXDC1 (maroon arrow) and incomplete duplication of LCR16a-021 NOMO1 (blue arrow)