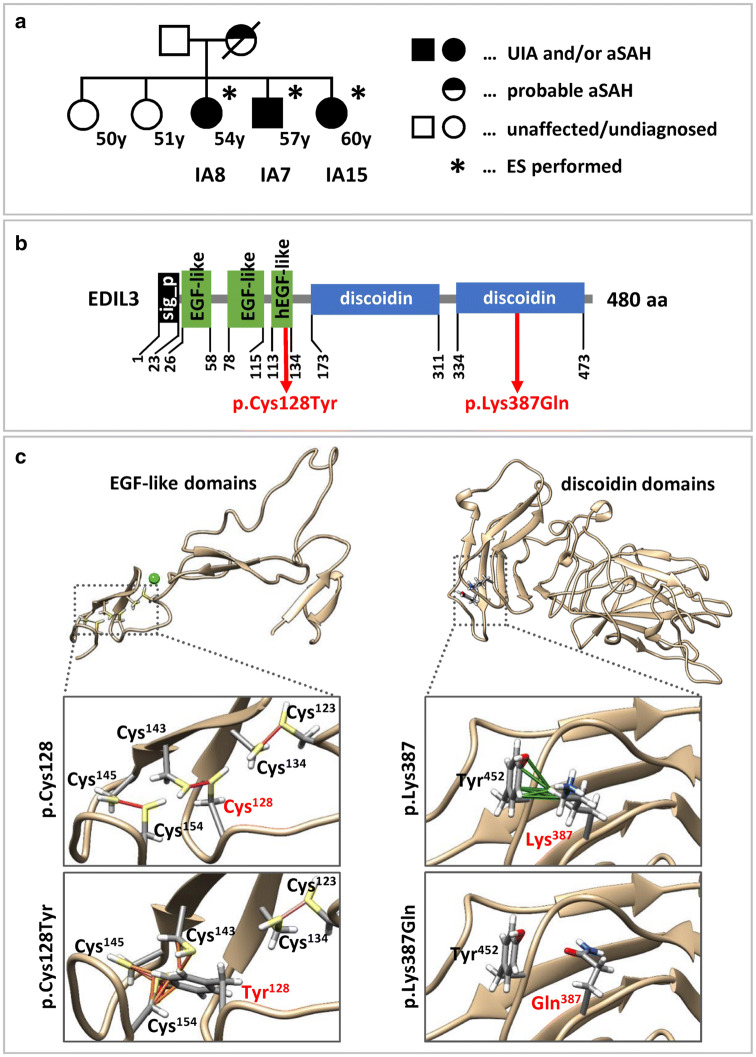
Fig. 1.
Pedigree for the affected family used for ES, domain structure of EDIL3 and structural impact of EDIL3 amino acid changes. a Pedigree. Exome sequenced individuals are marked with an asterisk. Criteria for defining the disease phenotypes (UIA and/or aSAH, probable aSAH) are outlined in the methods section. Age at inclusion is given in years (y). b Domain structure of EDIL3 comprising a signal peptide (sig_p, black), three (h)EGF-like domains (green) and 2 discoidin-like domains (blue) is shown relative to the size of the full-length protein. Localization of amino acid changes p.(Cys128Tyr) and p.(Lys387Gln) is indicated. aa, amino acid. c Structural impact of EDIL3 p.Cys128Tyr and p.Lys387Gln amino acid changes. Amino acids 128 and 387 and surrounding residues are shown as sticks; side chains are colored by element (hydrogen: white; carbon: grey; oxygen: red; nitrogen: blue; cysteine: yellow). Left column: (h)EGF-like domains of EDIL3 and the structural environment of Cys128. Ribbon representations show details of the hEGF-like domain with conserved cysteines that form three and two disulfide bridges (magenta lines) for wild-type (p.Cys128, top) and mutated (p.Cys128Tyr, bottom) EDIL3, respectively. In silico substitution of Cys128 for tyrosine resulted in loss of the disulfide bridge with Cys143 and overlaps of atomic Van-der Waals (VDW) spheres (orange lines) between Tyr128 with cysteines 143, 145 and 154 indicating possible non-covalent clashes. Right column: Discoidin domains of EDIL3 and the structural environment of Lys387. Ribbon representations show details of the discoidin domain for wild-type (p.Lys387, top) and mutated (p.Lys387Gln, bottom) EDIL3. For Lys387 six VDW contacts with carbons, a nitrogen and a hydrogen of the Tyr452 were identified. In silico substitution of Lys387 for glutamine resulted in loss of all VDW contacts with Tyr452. New VDW contacts or overlaps of Glu387 with adjacent residues were not predicted