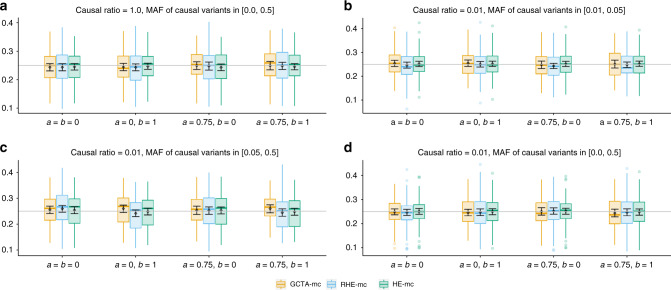
Fig. 2. Comparison of SNP heritability estimates from RHE-mc with GCTA-mc (GCTA with multiple variance components) and HE-mc (HE with multiple variance components) (N = 10,000 unrelated individuals, M = 593,300 array SNPs).
In a–d we compared heritability estimates from these methods under 16 different genetic architectures. We varied the MAF range of causal variants (MAF of CV), the coupling of MAF with effect size (a), and the effect of local LD on effect size (b = 0 indicates no dependence on LDAK weights and b = 1 indicates dependence on LDAK weights (see “Methods” section). We ran 100 replicates where the true heritability of the phenotype is 0.25. We run RHE-mc, HE-mc, and GCTA-mc using 24 bins formed by the combination of six bins based on MAF as well as four bins based on quartiles of the LDAK score of a SNP (see “Methods” section). Across all different genetic architectures, the relative biases range from −3.2% to 3.6% for RHE-mc, and from −3.2% to 5% for GCTA-mc, and from −2.6% to 1.45% for HE-mc. On average, RHE-mc has SEs that are 1.1 and 1.08 times larger than GCTA-mc and HE-mc, respectively. Black points and error bars represent the mean and ±2 SE. Each boxplot represents estimates from 100 simulations. Boxplot whiskers extend to the minimum and maximum estimates located within 1.5× interquartile range (IQR) from the first and third quartiles, respectively. The SEs are computed from 100 simulations (note that GCTA-mc did not run successfully on all 100 simulations).