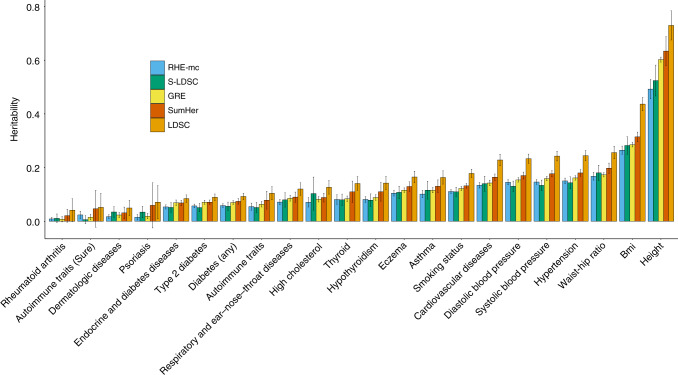
Fig. 4. Estimates of genome-wide SNP heritability from RHE-mc, LDSC, S-LDSC, GRE, and SumHer for 22 complex traits and diseases in the UK Biobank.
We restricted our analysis to N = 291,273 unrelated white British individuals. We applied all methods to M = 459,792 array SNPs (MAF > 1%). We ran S-LDSC with baseline-LD model. For every method, LD scores or LDAK weights are computed using in-sample LD among the SNPs, and we aim to estimate the SNP-heritability explained by the same set of SNPs. RHE-mc was applied to array SNPs with eight MAF/LD bins. Black error bars mark ±2 standard errors centered on the estimated heritability. We used a block Jackknife (number of blocks = 100) to estimate the standard errors. In Supplementary Fig. 7, we also report RHE-mc estimates of genome-wide SNP heritability on M = 4,824,392 imputed SNPs (MAF > 1%) with 8 MAF/LD bins and M = 7,774,235 imputed SNPs (MAF > 0.1%) with 144 MAF/LD bins (see “Methods” section).