Figure 1.
Generation of a GATA2VENUS Knockin Protein Reporter Mouse Line with Normal Hematopoiesis
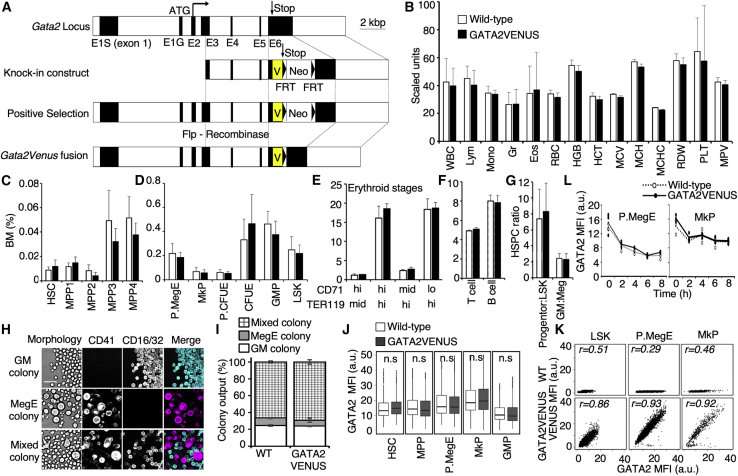
(A) Constructs used for GATA2VENUS knockin generation. The FRT PGK-Neo FRT was deleted by cross with a Flpe deleter mouse line. Black boxes indicate exons (also see Figure S1).
(B) Peripheral blood counts are not altered in GATA2VENUS mouse line. WBC, white blood cells (200 cells per mm3); Lym, percent lymphocytes of WBC (%); Mono, percent monocytes of WBC (0.1%); Gr, percent granulocytes of WBC (%); Eos, percent eosinophils of WBC (0.2%); RBC, red blood cells (2 ×105 cells per mm3); HGB, hemoglobin (0.2 g/dL); HCT, hematocrit (%); MCV, mean corpuscular volume (μm3); MCH, mean corpuscular hemoglobin (0.2 pg); MCHC, mean corpuscular hemoglobin concentration (g/dL); RDW, red cell distribution width (0.2%); PLT, platelets (104 per mm3); MPV, mean platelet volume (0.1 μm3) (n = 9 mice per genotype).
(C–G) Fusion of VENUS to GATA2 does not alter bone marrow composition. Data indicate bone marrow percentage of (C) HSCs and multipotent progenitors (n = 7 mice per genotype), (D) lineage committed progenitors (n = 10 mice per genotype), (E) early and late erythrocyte progenitors (n = 3 mice per genotype), (F) T and B cells (n = 3 mice per genotype), and (G) ratio of multipotent progenitors to lineage committed progenitors and granulocyte-monocyte progenitors to megakaryocyte-erythrocyte (MegE) progenitors (n = 10 mice per genotype).
(H and I) Colony-forming potential and output of HSCs is not altered in GATA2VENUS mouse line. (H) Single HSCs sorted into 384-wells in IMDM, FCS, BIT, SCF, EPO, TPO, IL-3, and IL-6. Granulocyte-monocyte (GM) colonies identified by morphology and FCγR expression. MegE colonies identified by morphology and CD41 expression. Scale bar, 50 μm. (I) Types of colonies formed from HSCs (n = 3 independent mice per genotype).
(J) Protein levels of GATA2 are not altered in different cell types in GATA2VENUS mouse line. Data were acquired using quantitative immunostaining against endogenous GATA2 protein. Data represented by box and whisker plots with median of GATA2 intensity (n = 3 independent mice per genotype).
(K) Endogenous GATA2 protein levels correlate to VENUS fusion levels. Data were acquired using quantitative immunostaining against GATA2 and VENUS and represented by a 2D plot of GATA2 and VENUS intensities. Number (r) in the plots indicate Pearson correlation coefficient (n = 3 independent mice per genotype).
(L) Normal stability of GATA2 fusion proteins in pre-MegE progenitors (preMegEs) (left panel) and megakaryocyte progenitors (MkPs) (right panel). Data were acquired using quantitative immunostaining against GATA2 in indicated cell types after treatment with 50 μM cycloheximide (protein translation inhibitor) and sampling of cells at the indicated time points (n = 3 independent mice per genotype).
Error bars in (B)–(G) and (I) = SD. Data in (J)–(L) indicate mean fluorescence intensity (MFI) of GATA2 and VENUS. Difference between wild-type and GATA2VENUS samples is non-significant unless specified. Two-sample t test; ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05.