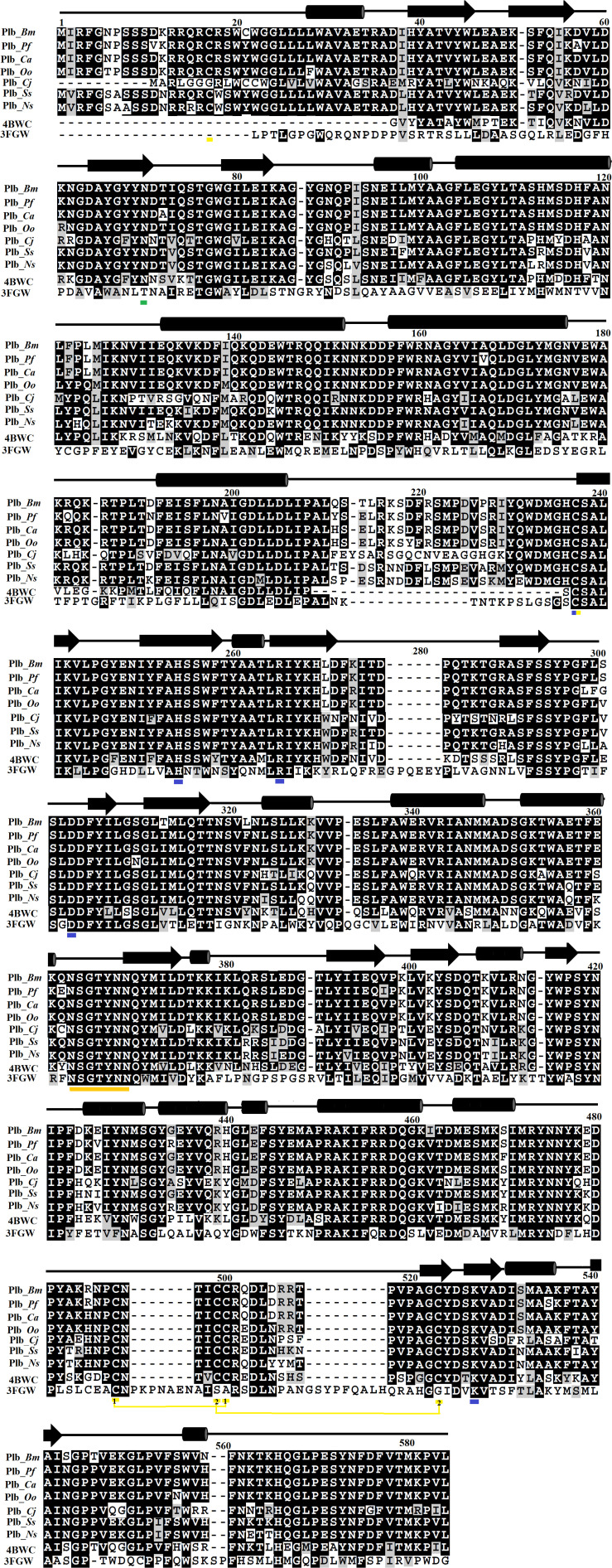
FIGURE 1.
Sequence alignment among snake venom phospholipases B (SVPLBs), PLB from bovine kidneys, and 66.3 kDa protein from Mus musculus. PLB_Bm, phospholipase B from Bothrops moojeni; PLB_Pf, phospholipase B from Protobothrops flavoviridis; PLB_Ca, phospholipase B from Crotalus atrox; PLB_Oo, phospholipase B from Ovophis okinavensis; PLB_Cj, phospholipase B from Coturnix japonica; PLB_Ss, phospholipase B from Spilotes sulphureus; PLB_Ns, phospholipase B from Notechis scutatus; 4BWC, phospholipase B like protein 1 from bovine kidneys; 3FGW, 66.3 kDa protein from M. musculus. The amino acid residues involved in catalysis are underlined with blue, and the cysteine residues are underlined with yellow. The consensus lipase sequence is underlined with gold color. The cysteine residues that make disulfide bridges are linked (yellow lines). The putative N-glycosylation amino acid residues are underlined in green. The secondary structure elements (α-helices and β-strands) are shown above the sequence.