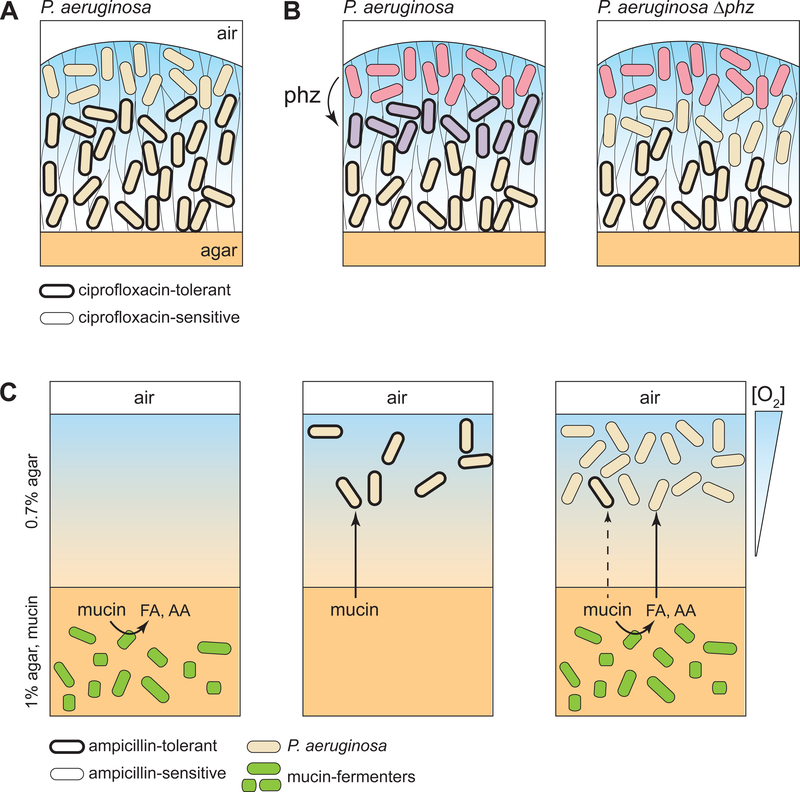
Figure 4. Implications of metabolic heterogeneity for antibiotic tolerance of P. aeruginosa populations.
Three studies addressing the effects of metabolic heterogeneity on antibiotic tolerance are represented. (A, B) Tolerance to ciprofloxacin in P. aeruginosa colony biofilms. (A) Williamson et al. used selective GFP labeling of metabolically active or inactive cells, followed by antibiotic exposure, FACS, and CFU counting, to show that the air-exposed metabolically active subpopulation is more susceptible to antibiotics [67]. (B) Schiessl et al. used SRS microscopy to show that phenazine-producing biofilms contain two bands of metabolically active cells (red and purple), while phenazine-null biofilms contain one pronounced band of metabolically active cells (red) [68]. Based on CFU counts, phenazine-producing biofilms show less antibiotic susceptibility, suggesting that the lower band of metabolic activity in these biofilms contains cells in a differentiated state with higher antibiotic tolerance. (C) An anaerobic community of mucin fermenters embedded in agar produces short-chain fatty acids (FA) and amino acids (AA). These products can serve as carbon sources and support replication of a P. aeruginosa population present in the air-exposed portion of the agar block. Interestingly, while the fermenters are sensitive to ampicillin in mono-and co-culture (left and right panels, respectively), P. aeruginosa is only sensitive in co-culture [78, 79].