Figure 1. Data-Driven Phenotyping Using Subgraph Augmented Nonnegative Matrix Factorization.
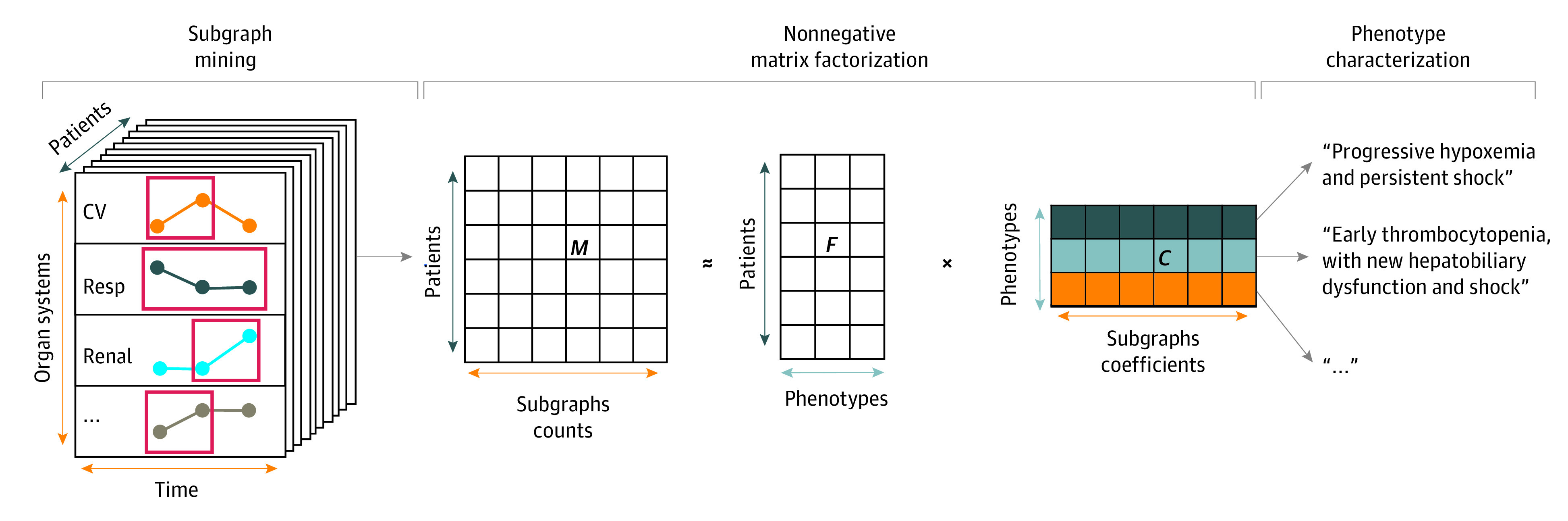
Subgraph mining extracts representative subgraphs from the trajectories of the 6 pediatric Sequential Organ Failure Assessment subscores in patients with MODS, which results in a matrix, M, of patient-subgraph counts. Nonnegative matrix factorization is used to derive a matrix, F, of hidden features (in this case the phenotypes), and a matrix of mixture coefficient C with the coefficients that compose the subgraph-based phenotypes. The factorization is done by iteratively updating F and C using the sparse nonnegative matrix factorization with sparseness in the left factor algorithm to gradually reduce the error between M and F × C. Once the nonnegative matrix factorization is completed, the final phenotypes can be characterized given the highly interpretable nature of the algorithm. Of note, the size of the matrices, number of groups, and the phenotype characterization examples here are used for illustration purposes only. CV indicates cardiovascular; resp, respiratory.
