Abstract
Long non-coding RNAs (lncRNAs) are newly introduced as tumor-related molecules. They are being considered as potential diagnostic and therapeutic targets in cancer studies. Here, we have assessed the importance of CYTOR (linc00152) as a biomarker for the detection of breast cancer in both tumoral tissue and plasma. The relative expression of breast cancer-associated CYTOR was measured in 20 tumoral and paired margin tissues, and moreover, in 80 plasma samples by real-time-polymerase chain reaction. In addition, plasma levels of CA 15-3 were measured using the ELISA test. The receiver operating characteristic curve (ROC) was applied for assessing the sensitivity and specificity of tested biomarkers. The results of the present study disclosed significantly increases in the CYTOR expression levels in tumor tissue with relative quantitation (RQ) equals to 5.15 (P<0.001) and in plasma (RQ =3.03, P<0.01) of breast cancer patients. The area under the ROC curve (AUC) of circulating CYTOR was 0.907 (95% CI: 0.842-0.972, P<0.001), while it was 0.868 (95% CI: 0.783–0.952, P<0.001) for CA 15-3. Based on these findings, we suggest lncRNA CYTOR as a new potential biomarker for breast cancer detection in both solid tumor tissue and plasma.
Key Words: Breast cancer, lncRNA, CYTOR, biomarker
Breast cancer (BC) is one of the primary health problems worldwide (1, 2). According to the report of Non-Eommunicable Diseases Research Center of Iran, BC is the most prevalent cancer among the women in Kermanshah, West of Iran (33.02 new cases per 100000 women, in 2016) (3).
Early detection is critical for reducing the mortality and improving prognosis of this malignancy. Blood-based testing is recom-mended for detecting cancer biomarkers due to its easiness and lower invasiveness (4). However, measuring conventional serum biomarkers for BC detection, such as carbohydrate antigens (CA125 and CA15-3) and carcinoembryonic antigen (CEA) are not convincing in early detection (5). For this reason, new biomarker detection seems to be necessary for early diagnosis and monitoring of BC.
By definition, long non-coding RNAs (lncRNAs) are biomolecules which their length is longer than 200 nucleotides. They can act as oncogenes or tumor suppressor genes, and their aberrant expression is probably involved in the progression of tumors (6). Linc00152, known as a cytoskeleton regulator long non-coding RNA (CYTOR), is an 828 bp gene located on the human chromosome 2p11.2 (7). CYTOR is up-regulated in various cancer types, including all subtypes of BC (8). Furthermore, recently it has been indicated that CYTOR could act as a circulating biomarker for the esophageal squamous-cell carcinoma, gastric cancer, and non-small-cell lung cancer (9-11). However, the potential clinic value of circulating CYTOR (c.CYTOR) in BC has not been investigated. Therefore, the aim of the present investigation was to study the levels of c.CYTOR in plasma of BC patientsto show its role as a novel BC biomarker.
Materials and methods
Participants and sample collection
The BC cases were recruited at the time of the diagnosis, between 2016 and 2017 at the Bistoon Hospital, Kermanshah Province, Iran. The mean age of patients was about 49 years (range 30–68 years), and the mean age of healthy controls was 48 years (range 26–64 years, P = 0.14). The study was approved by the Ethics Committee of Kermanshah University of Medical Sciences and informed consent was obtained from all the subjects. Only new confirmed BC cases without a previous diagnosis of any malignant disease and the manifestation of any acute disease or injury were included in the present study. The first group consisted of tumoral and paired adjacent non-tumoral frozen tissues from 20 BC cases at the time of diagnosis. The second group consisted of 40 BC patients who were recruited only for peripheral blood sampling during diagnosis (before surgery and any therapy). Besides, a control group included 40 healthy blood donors. All participants were women with a Kurdish ethnic background from Kermanshah Province, Iran.
Plasma samples collection
The whole peripheral blood samples were collected in 5 mL EDTA containing tubes, then centrifuged (3500×g for 20 min) within an hour after blood collection to achieve plasma separation. The cellular fraction of prepared blood was separated, and plasma samples were stored at −80 °C.
RNA extraction and complementary DNA (cDNA) synthesis
Fresh frozen tissues were powdered in liquid nitrogen, and then total RNA was extracted by Qizol (QIAGEN, Germany) according to the manufacturer’s procedure. In addition, total RNA was isolated from 200 μL of plasma samples by using miRNeasy Plasma/Serum Kit (QIAGEN, Germany), and it was eluted into 14 μL of elution solution. The concentration and purity of the extracted RNA were evaluated by the Nanodrop spectrop-hotometer (Thermo Scientific, Waltham, MA). Complementary DNA (cDNA) was synthesized from the isolated total RNA by applying a QuantiTect Reverse Transcription Kit (QIA-GEN, Germany).
Quantitative real-time PCR (qRT-PCR)
The sequences of selected primers used for qRT-PCR were 5’CTGGATGGTCGCTGCTTT-TT3’ (sense) and 5’GATCTGAAGACAGGC-ACGGG3’ (antisense) for CYTOR (8), and 5’CTCGCTTCGGCAGCACA3’ (sense) and 5’ AACGCTTCACGAATTTGCGT3’ (antisense) for U6-snRNA (12) as an internal control. The qRT-PCR assay was performed using the Rotor-gene (Corbett Research, Germany) with SYBR Premix Ex Taq II (Takara, China). The PCR reaction conditions were initial denaturation at 95 °C for 30 s, followed by 45 PCR cycles at 95 °C for 10 s, 60 °C for 15 s and 72 °C for 30 s. We used the 2−△△Ct method to determine the relative quantitation (RQ) in the cancerous samples relative to the control samples.
Measurement of CA 15-3 in plasma
The CA15-3 was assessed in accordance with the manufacturer’s protocol and reference intervals. This conventional tumor marker was measured in 80 plasma samples (40 healthy controls and 40 BC patients) using an ELISA kit (Monobind, USA). The CA15-3 threshold value provided by the company was equal to 37 U/ml. For plasma tumor marker definition, when its level exceeded a threshold value, the samples were considered to be positive.
Statistical analysis
Comparisons between groups were analyzed with Student’s t-test and X2 tests. Statistical analyses were performed using SPSS (v.16.0) software. A P-value less than 0.05 was considered significant. Receiver operating characteristic (ROC) curves and the area under the curve (AUC) were used to assess the feasibility of using plasma CYTOR as a diagnostic tool for BC detection.
Results
The results of immunohistochemical analysis for hormone-responsiveness markers among 40 BC patients revealed 31 (77.5%) cases with estrogen (ER)-positive, 28 (70%) cases with progesterone (PR)-positive, 19 (47.5%) cases with HER2-positive, and 40 (100%) cases with Ki67-positive markers. Moreover, pathological examination reports of BC samples indicated the frequencies of 27.5%, 47.5%, and 25%, for stages I, II, and III, respectively.
The expression level of CYTOR was significantly higher in tumoral tissues
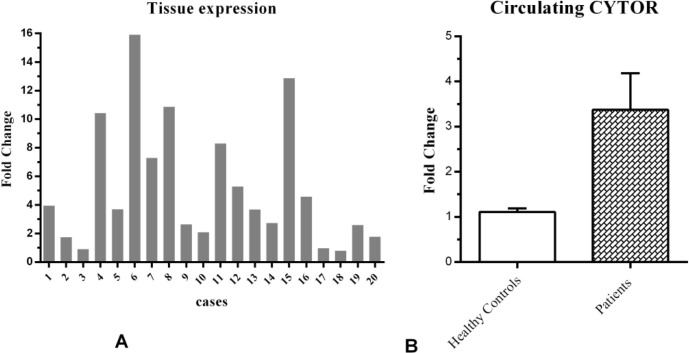
Using qRT-PCR, the relative expression level of CYTOR was measured in 20 breast tumor tissues and their adjacent non-tumoral tissues. The level of CYTOR gene expression in tumor tissues was significantly higher in comparison with paired adjacent non-tumoral tissues (P<0.001, Figure 1A).
Fig.1.
Relative expression of CYTOR . A: CYTOR expression in 20 paired breast tumor tissues and their adjacent non-tumoral tissues; B: relative expression levels of CYTOR in plasma of breast cancer patients in comparison with healthy controls
The average RQ of tumor tissues in comparison with adjacent non-tumoral tissues was 5.15. There was no significant correlation between the tumoral tissue expression of CYTOR and the status of hormone-responsiveness markers (ER, PR, or HER2) or tumor stages.
The plasma levels of c.CYTOR was signific-antly higher in BC patients
The relative level of circulating CYTOR expression was determined in each plasma sample. We found that the expression level of CYTOR in plasma samples collected from BC patients was significantly higher than the cancer-free donors (RQ= 3.03, P<0.01, Figure 1B). This excess lncRNA CYTOR in the patient's plasma is probably released from the tumor. Also, the analysis showed the absence of a significant correlation between the c.CYTOR levels and the status of hormone-responsiveness markers (ER, PR, or HER2) or tumor stages.
Analysis of c.CYTOR stability in plasma
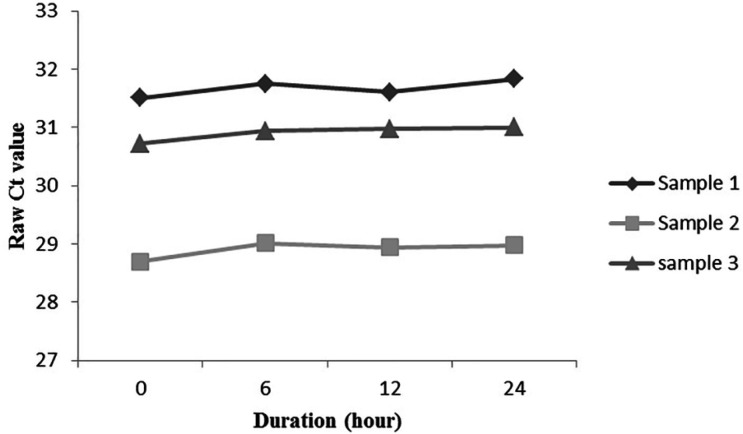
To analyze the lncRNA stability in plasma, three plasma samples were divided into 4 equal parts, and incubated at room temperature for 0, 6, 12, and 24 h. Through defined time, no considerable variation was observed in plasma CYTOR levels (P<0.05, Figure 2). These results indicated that CYTOR remained stable in plasma for at least 24 h, even at room temperature.
Fig.2.
Stability assessment of circulating CYTOR in plasma. Three plasma samples were assessed for CYTOR stability right after sampling (0 h), after 6, 12, and 24 h incubation of plasma at room temperature. Ct: cycle threshold
Diagnostic value of c.CYTOR and CA 15-3 for BC
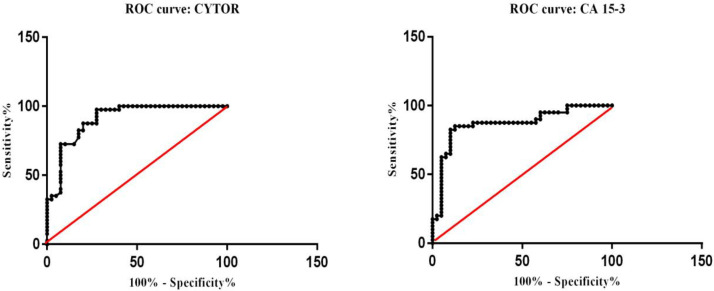
To evaluate the potential clinical applic-ation of c.CYTOR, the ROC curve was constructed according to 80 BC patients and cancer-free controls data. The AUC of CYTOR was 0.907 (95% CI: 0.842-0.972, P<0.001, Figure 3A) for distinguishing BC patients from controls. Furthermore, for the comparison of the diagnostic value of c.CYTOR with a conventional plasma biomarker, cancer antigen 15-3 (CA 15-3) levels were assessed in 80 plasma samples (AUC= 0.868, 95% CI: 0.783–0.952, P<0.001, Figure 3B)
Fig.3.
ROC curve alnaysis of CYTOR and CA 15-3 in plasma of breast cancer patients. A: evaluation of the diagnostic value of circulating CYTOR; B: CA 15-3 diagnostic value
Discussion
Regarding the stability of circulating lncRNAs in plasma/serum, and their transcripts as the final functional products, assessing the expression levels of a lncRNA could directly show the levels of an active molecule. Accordingly, lncRNA transcripts have a potential value as a candidate for a non-invasive clinical index (13).
The present study was designed to assess the expression pattern and diagnostic value of lncRNA CYTOR in BC. First, we investigated the expression levels of CYTOR in BC tissues in comparison with adjacent non-tumoral tissues. We showed that CYTOR levels were higher in breast tumors in comparison with adjacent non-tumoral tissues. These results were in accordance with the previous studies, which suggested that CYTOR may play an oncogenic role in BC (8, 14-16).
Since it can be speculated that lncRNAs in circulation are unstable, thus, we then attempted to assess the stability of CYTOR in circulation. The results disclosed that CYTOR levels remained stable in plasma, which was consistent with previous studies (9, 11). Afterwards, we further found that c.CYTOR levels were significantly higher in the plasma of BC patients in comparison with healthy individuals, suggesting that CYTOR could offer a relatively high diagnostic efficacy for BC. The AUC of ROC curve for c.CYTOR was higher than CA 15-3 which is one of the conventionally used BC biomarkers (17, 18).
From eight reported articles related to the expression of c.CYTOR in plasma/serum, only one reported no significant differences between gastric cancer patients and normal controls (19), while others reported the upregulation of c.CYTOR in gastric cancer (10, 20, 21), hepatocellular carcinoma (22, 23), esophageal squamous-cell carcinoma (9), and non-small cell lung cancer (11). The results of our study, for the first time, suggested that CYTOR could be a circulating biomarker for BC diagnosis.
CYTOR has been introduced as an oncogenic lncRNA in different types of cancers, including tongue squamous cell carcinoma, pancreatic cancer, lung cancer, gastric cancer, liver cancer, and BC (16, 24-28). The molecular mechanism of CYTOR oncogenesis is not well understood, yet. It is shown that Yin Yang 1 (YY1), as a transcription factor, binds to the promoter of CYTOR and suppresses its transcription; while transcription Factor 3 (TCF3) could activate the transcription of CYTOR (16, 29). On the other hand, it was demonstrated that CYTOR overexpression promotes tumor cell cycle progression, cell viability, invasion, and epidermal-mesenchymal transition (EMT) (7, 8, 14, 30, 31).
Wu et al. (2018) showed that CYTOR knockdown promotes BRCA1/ phosphatase and tensin homolog (PTEN), ) expression through DNA methyltransferases (14). While Shen et al. (2018) demonstrated that knockdown or overexpression of CYTOR had no significant influence on the mRNA level of PTEN; instead, CYTOR impairs PTEN protein stability through triggering ubiquitination and degradation of PTEN protein by E3 ligase neural precursor cell expressed developmentally downregulated protein 4-1 (NEDD4-1) (16). PTEN acts via its phosphatase protein, which is involved in the cell cycle regulation and prevention of uncontrolled cell division (32).
CYTOR through the epidermal growth factor receptor (EGFR)-dependent pathway could promote cell viability (33) and through binding to the enhancer of zeste homolog 2 (EZH2) and consequently repressing p15 and p21 triggers cell cycle progression in gastric tumor (34). Furthermore, CYTOR through EGFR/PI3K/ATK pathway was shown to support cell viability and invasion in lung cancer (35). Some studies indicated that CYTOR can promote EMT in gastric cancer cells, liver cancer cells, and BC cells (28, 30, 36). EMT is a cellular process that is involved in tumor metastasis. In liver cancer cells, CYTOR could promote EMT by interacting with EZH2 and suppression of E-cadherin (28).
Totally, it seems that CYTOR through activation of EGFR signaling and suppression of PTEN and E-cadherin promotes EMT, which plays an important role in BC progression.
In conclusion, the findings of the present study suggest a role for plasma levels of lncRNA CYTOR as a candidate biomarker for BC that its level significantly increases in plasma of patients affected with BC. However, the biomarker role of this lncRNA needs to be investigated in other populations.
Acknowledgments
This study was funded by Kermanshah University of Medical Sciences, Iran (Grant number: 95680).
Conflict of interest
Authors declare no conflict of interest in this study.
References
- 1.Yari K, Rahimi Z, Payandeh M, et al. MMP-7 A-181G Polymorphism in Breast Cancer Patients from Western Iran. Breast Care (Basel) 2015;10:398–402. doi: 10.1159/000442231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharifian A, Pourhoseingholi MA, Emadedin M, et al. Burden of Breast Cancer in Iranian Women is Increasing. Asian Pac J Cancer Prev. 2015;16:5049–52. doi: 10.7314/apjcp.2015.16.12.5049. [DOI] [PubMed] [Google Scholar]
- 3.Farzadfar F. National and Sub-National Burden of Diseases’ Atlas Islamic Republic of Iran 1990-2016. 2016. vailable from: https://vizit.report/panel/cancer/en/main.html.
- 4.Elmore JG, Armstrong K, Lehman CD, et al. Screening for breast cancer. JAMA. 2005;293:1245–56. doi: 10.1001/jama.293.10.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maric P, Ozretic P, Levanat S, et al. Tumor markers in breast cancer--evaluation of their clinical usefulness. Coll Antropol. 2011;35:241–7. [PubMed] [Google Scholar]
- 6.Qiu MT, Hu JW, Yin R, et al. Long noncoding RNA: an emerging paradigm of cancer research. Tumour Biol. 2013;34:613–20. doi: 10.1007/s13277-013-0658-6. [DOI] [PubMed] [Google Scholar]
- 7.Notzold L, Frank L, Gandhi M, et al. The long non-coding RNA LINC00152 is essential for cell cycle progression through mitosis in HeLa cells. Sci Rep. 2017;7:2265. doi: 10.1038/s41598-017-02357-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Grembergen O, Bizet M, de Bony EJ, et al. Portraying breast cancers with long noncoding RNAs. Sci Adv. 2016;2:e1600220. doi: 10.1126/sciadv.1600220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu HB, Jie HY, Zheng XX. Three Circulating LncRNA Predict Early Progress of Esophageal Squamous Cell Carcinoma. Cell Physiol Biochem. 2016;40:117–25. doi: 10.1159/000452529. [DOI] [PubMed] [Google Scholar]
- 10.Li Q, Shao Y, Zhang X, et al. Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumour Biol. 2015;36:2007–12. doi: 10.1007/s13277-014-2807-y. [DOI] [PubMed] [Google Scholar]
- 11.Li N, Feng XB, Tan Q, et al. Identification of Circulating Long Noncoding RNA Linc00152 as a Novel Biomarker for Diagnosis and Monitoring of Non-Small-Cell Lung Cancer. Dis Markers. 2017;2017:7439698. doi: 10.1155/2017/7439698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H, Chen F, He Y, et al. Sensitivity of non-small cell lung cancer to erlotinib is regulated by the Notch/miR-223/FBXW7 pathway. Biosci Rep. 2017:37. doi: 10.1042/BSR20160478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu N, Chen F, Wang F, et al. Clinical significance of high expression of circulating serum lncRNA RP11-445H224 in breast cancer patients: a Chinese population-based study. Tumour Biol. 2015;36:7659–65. doi: 10.1007/s13277-015-3469-0. [DOI] [PubMed] [Google Scholar]
- 14.Wu J, Shuang Z, Zhao J, et al. Linc00152 promotes tumorig--enesis by regulating DNMTs in triple-negative breast cancer. Biomed Pharmacother. 2018;97:1275–81. doi: 10.1016/j.biopha.2017.11.055. [DOI] [PubMed] [Google Scholar]
- 15.He Y, Hu H, Wang Y, et al. ALKBH5 Inhibits Pancreatic Cancer Motility by Decreasing Long Non-Coding RNA KCNK15-AS1 Methylation. Cell Physiol Biochem. 2018;48:838–46. doi: 10.1159/000491915. [DOI] [PubMed] [Google Scholar]
- 16.Shen X, Zhong J, Yu P, et al. YY1-regulated LINC00152 promotes triple negative breast cancer progression by affecting on stability of PTEN protein. Biochem Biophys Res Commun. 2019;509:448–54. doi: 10.1016/j.bbrc.2018.12.074. [DOI] [PubMed] [Google Scholar]
- 17.Safi F, Kohler I, Rottinger E, et al. The value of the tumor marker CA 15-3 in diagnosing and monitoring breast cancer A comparative study with carcinoembryonic antigen. Cancer. 1991;68:574–82. doi: 10.1002/1097-0142(19910801)68:3<574::aid-cncr2820680322>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 18.Duffy MJ, Evoy D, McDermott EW. CA 15-3: uses and limitation as a biomarker for breast cancer. Clin Chim Acta. 2010;411:1869–74. doi: 10.1016/j.cca.2010.08.039. [DOI] [PubMed] [Google Scholar]
- 19.Xian HP, Zhuo ZL, Sun YJ, et al. Circulating long non-coding RNAs HULC and ZNFX1-AS1 are potential biomarkers in patients with gastric cancer. Oncol Lett. 2018;16:4689–98. doi: 10.3892/ol.2018.9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang T, Zeng H, Chen W, et al. Helicobacter pylori infection, H19 and LINC00152 expression in serum and risk of gastric cancer in a Chinese population. Cancer Epidemiol. 2016;44:147–53. doi: 10.1016/j.canep.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 21.Mao Y, Tie Y, Du J, et al. LINC00152 promotes the proliferation of gastric cancer cells by regulating B-cell lymphoma-2. J Cell Biochem. 2019;120:3747–56. doi: 10.1002/jcb.27655. [DOI] [PubMed] [Google Scholar]
- 22.Li J, Wang X, Tang J, et al. HULC and Linc00152 Act as Novel Biomarkers in Predicting Diagnosis of Hepatocellular Carcinoma. Cell Physiol Biochem. 2015;37:687–96. doi: 10.1159/000430387. [DOI] [PubMed] [Google Scholar]
- 23.Yuan W, Sun Y, Liu L, et al. Circulating LncRNAs Serve as Diagnostic Markers for Hepatocellular Carcinoma. Cell Physiol Biochem. 2017;44:125–32. doi: 10.1159/000484589. [DOI] [PubMed] [Google Scholar]
- 24.Yu J, Liu Y, Guo C, et al. Upregulated long non-coding RNA LINC00152 expression is associated with progression and poor prognosis of tongue squamous cell carcinoma. J Cancer. 2017;8:523–30. doi: 10.7150/jca.17510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu X, Lin Y, Sui W, et al. Analysis of distinct long noncoding RNA transcriptional fingerprints in pancreatic ductal adenocarcinoma. Cancer Med. 2017;6:673–80. doi: 10.1002/cam4.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang PP, Wang YQ, Weng WW, et al. Linc00152 promotes Cancer Cell Proliferation and Invasion and Predicts Poor Prognosis in Lung adenocarcinoma. J Cancer. 2017;8:2042–50. doi: 10.7150/jca.18852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia T, Liao Q, Jiang X, et al. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci Rep. 2014;4:6088. doi: 10.1038/srep06088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng X, Zhao XF, Liang XQ, et al. Linc00152 promotes cancer progression in hepatitis B virus-associated hepatocellular carcinoma. Biomed Pharmacother. 2017;90:100–8. doi: 10.1016/j.biopha.2017.03.031. [DOI] [PubMed] [Google Scholar]
- 29.Zheng L, Hu N, Zhou X. TCF3-activated LINC00152 exerts oncogenic role in osteosarcoma through regulating miR-1182/CDK14 axis. Pathol Res Pract. 2019;215:373–80. doi: 10.1016/j.prp.2018.12.031. [DOI] [PubMed] [Google Scholar]
- 30.Hu XL, Wang J, He W, et al. Down-regulation of lncRNA Linc00152 suppressed cell viability, invasion, migration, and epithelial to mesenchymal transition, and reversed chemo-resistance in breast cancer cells. Eur Rev Med Pharmacol Sci. 2018;22:3074–84. doi: 10.26355/eurrev_201805_15067. [DOI] [PubMed] [Google Scholar]
- 31.Xu S, Kong D, Chen Q, et al. Oncogenic long noncoding RNA landscape in breast cancer. Mol Cancer. 2017;16:129. doi: 10.1186/s12943-017-0696-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yari K, Payandeh M, Rahimi Z. Association of the hypermethylation status of PTEN tumor suppressor gene with the risk of breast cancer among Kurdish population from Western Iran. Tumour Biol. 2016;37:8145–52. doi: 10.1007/s13277-015-4731-1. [DOI] [PubMed] [Google Scholar]
- 33.Zhou J, Zhi X, Wang L, et al. Linc00152 promotes proliferation in gastric cancer through the EGFR-dependent pathway. J Exp Clin Cancer Res. 2015;34:135. doi: 10.1186/s13046-015-0250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen WM, Huang MD, Sun DP, et al. Long intergenic non-coding RNA 00152 promotes tumor cell cycle progression by binding to EZH2 and repressing p15 and p21 in gastric cancer. Oncotarget. 2016;7:9773–87. doi: 10.18632/oncotarget.6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Xiang C, Wang Y, et al. lncRNA LINC00152 knockdown had effects to suppress biological activity of lung cancer via EGFR/PI3K/AKT pathway. Biomed Pharmacother. 2017;94:644–51. doi: 10.1016/j.biopha.2017.07.120. [DOI] [PubMed] [Google Scholar]
- 36.Zhao J, Liu Y, Zhang W, et al. Long non-coding RNA Linc00152 is involved in cell cycle arrest, apoptosis, epithelial to mesenchymal transition, cell migration and invasion in gastric cancer. Cell Cycle. 2015;14:3112–23. doi: 10.1080/15384101.2015.1078034. [DOI] [PMC free article] [PubMed] [Google Scholar]