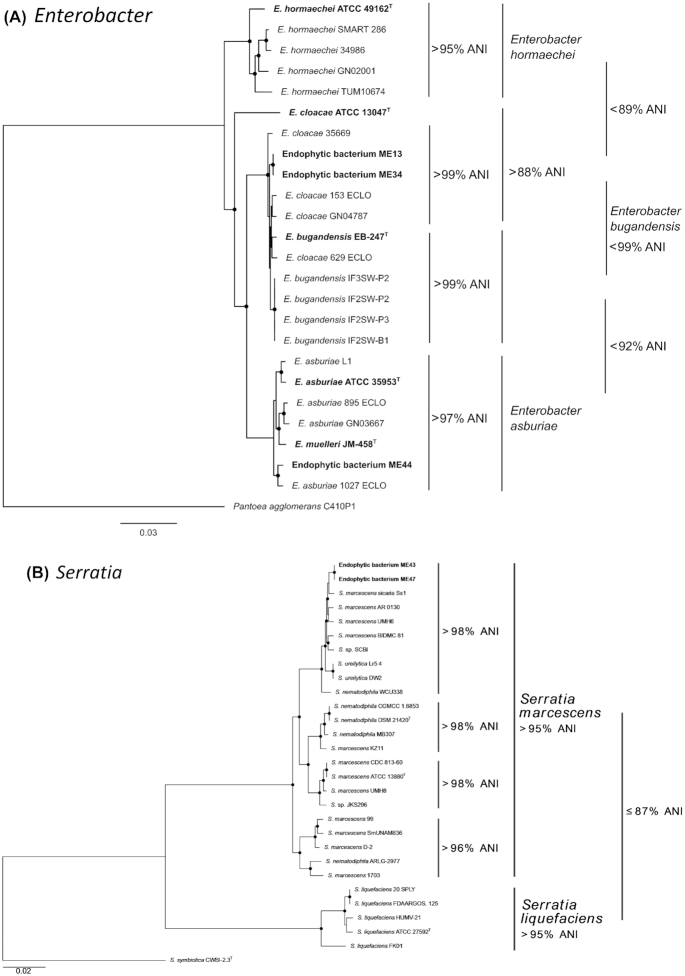
Figure 4.
Average nucleotide identity (ANI) and core genome analysis of medicinal plant endophytes (ME) belonging to the order Enterobacterales: (A)Enterobacter and (B)Serratia. (A) Core-gene phylogeny of ME belonging to Enterobacter species. A 1912 core-gene alignment generated by Roary was used to construct a maximum likelihood tree highlighting the placement of Enterobacter endophytes. Isolates ME13 and ME34 placed within the E. cloacae species clade, whilst ME44 placed within the E. asburiae clade. (B) Core-gene phylogeny of ME belonging to Serratia species. A 255 core-gene alignment generated by Roary was used to construct a maximum likelihood phylogeny highlighting the placement of isolated Serratia endophytic bacteria. ME43 and ME47 were placed within the Serratia marcescens species clade with >95% ANI to other members of the species. Phylogenetic trees were constructed with GTR model with gamma substitution and supported by 100 bootstraps. Nodes with black circles represent >90% bootstrap support. Scale bar = substitutions per site.