Fig 3. Evaluation of Maker2 protein predictions.
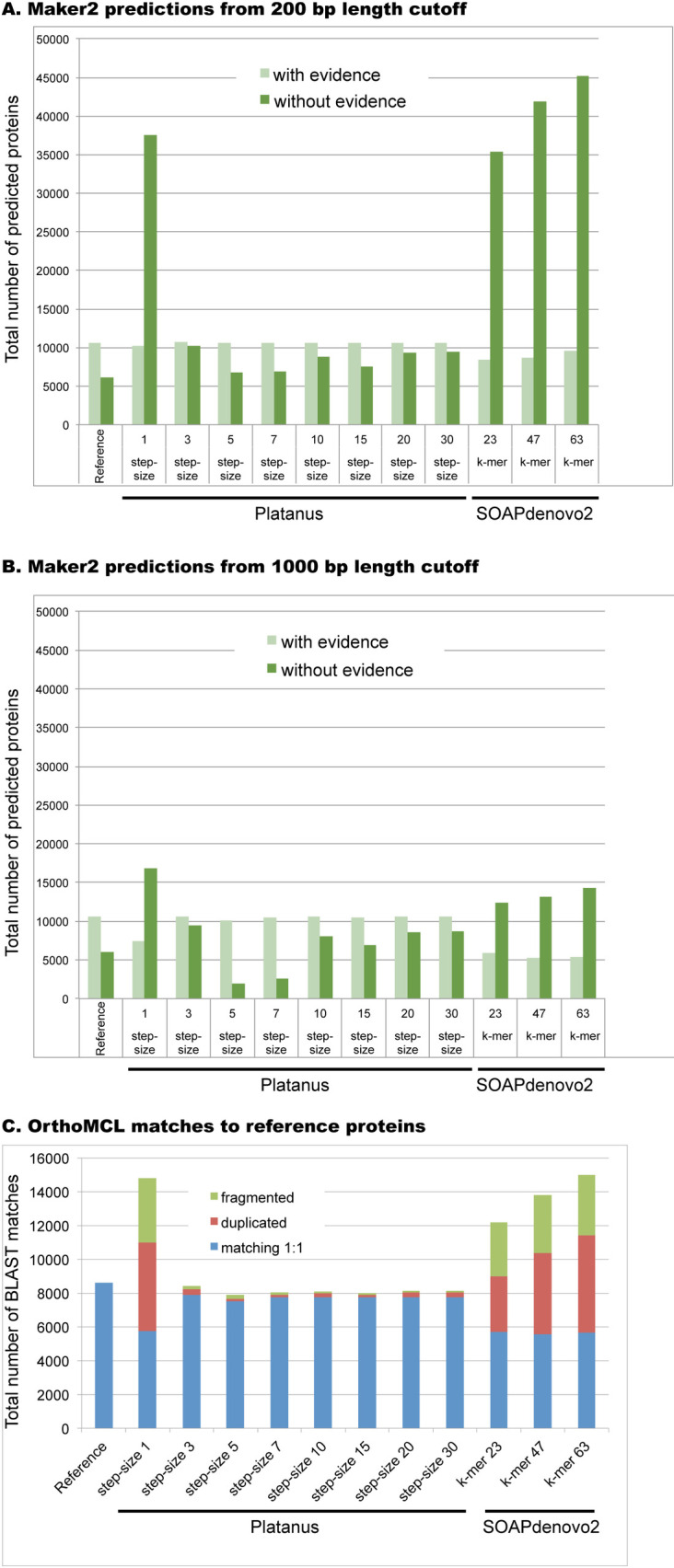
(A) Predicted proteins generated from assemblies with 200 bp size cutoff. (B) Predicted proteins generated from assemblies with 1000 bp size cutoff. For both A and B we have indicated the two types of protein predictions from Maker2: homology evidence-supported (light green) and those without evidentiary support (dark green). (C) Fragmentation and duplication from OrthoMCL-generated blast matches of each assembly (200bp cutoff; all proteins combined) relative to reference proteins. For each assembly, we were able to quantify 1:1 matches (blue), cases of fragmentation (non-overlapping matches to a single reference protein, green), and duplication (multiple overlapping matches to a single reference protein, red). To screen out paralogous protein families, we ignored all cases where the reference assembly provided a non-self match (see Methods); thus the number of protein matches is substantially lower than the total number of predicted proteins for each assembly.
