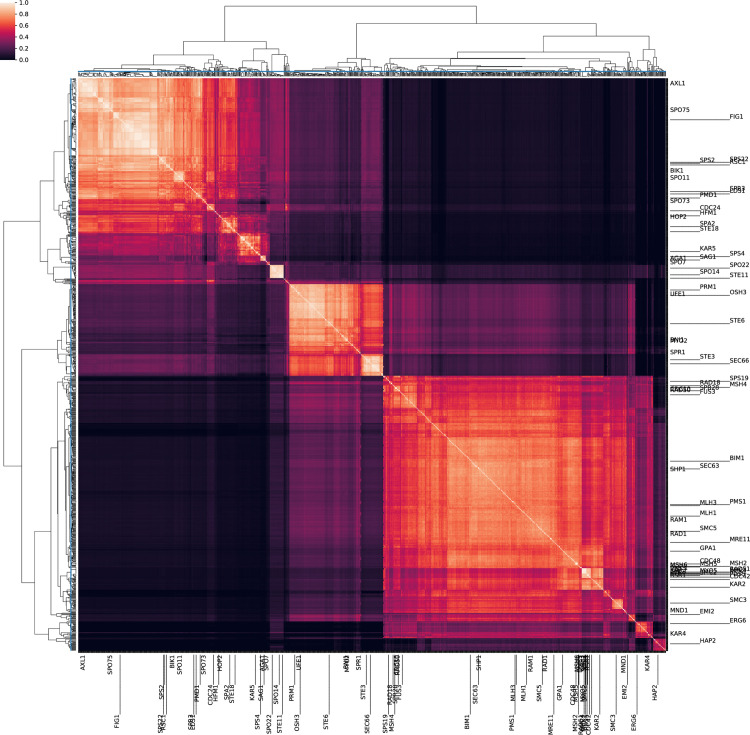
Fig 5. HogProf’s reproductive network.
A list of proteins known to be involved in sexual reproduction was compiled and mapped to OMA HOGs. These queries were used to search for the 20 closest coevolving HOGs in an LSH forest containing all HOGs in OMA. A Jaccard kernel was generated by performing an All vs All comparison of the Hash signatures of search results and queries. UPGMA clustering was performed on the rows and columns of the kernel. A cutoff distance of .995 (blue lines) was manually chosen to limit cluster sizes to less than 50 HOGs. This generated a total of 215 clusters of HOGs. Names for queries are shown with Saccharomyces cerevisiae gene names (apart from Hap2 which is not present in fungi).