Fig. 1. Thor-HyperTRIBE identifies specific RNAs as 4E-BP targets in Drosophila S2 cells.
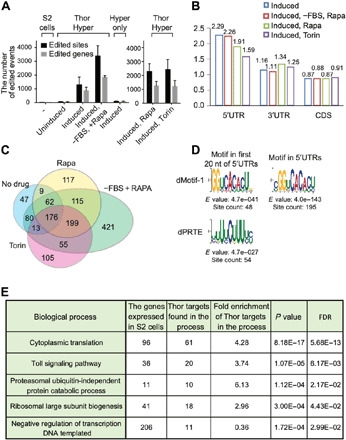
(A) Thor-HyperTRIBE but not hyper-ADARcd alone (Hyper only) edits transcripts after copper induction. The editing sites (black bars) and editing genes (gray bars) in Thor-HyperTRIBE significantly increased in rapamycin- or Torin-1–treated cells and more than doubled in rapamycin-treated plus serum-deprived cells. The number of editing events in cells expressing hyper-ADARcd alone is comparable to that of control S2 cells (N = 2, +SEM). (B) Editing sites identified by Thor-HyperTRIBE are enriched in 5′UTR of mRNAs. (C) Venn diagram of Thor-HyperTRIBE target RNAs reproducibly identified under different conditions shows that the targets are consistent, although significantly more were identified with serum deprivation and rapamycin treatment combined. The transcripts edited in S2 or Hyper only control were removed from the target list. One hundred seventy-six target genes were identified in all conditions. (D) Consensus motifs from the 5′UTRs of 968 Thor-HyperTRIBE targets (listed in table S1), which were reproducibly detected in rapamycin or Torin-1 treatment condition. The GGUCACACU motif is identified in both cases with 195 counts (~20%) in the entire 5′UTR of the targets. (E) Table of enriched GO term biological processes in 968 Thor-HyperTRIBE targets reproducibly detected in rapamycin or Torin-1 treatment condition. FDR, false discovery rate.
