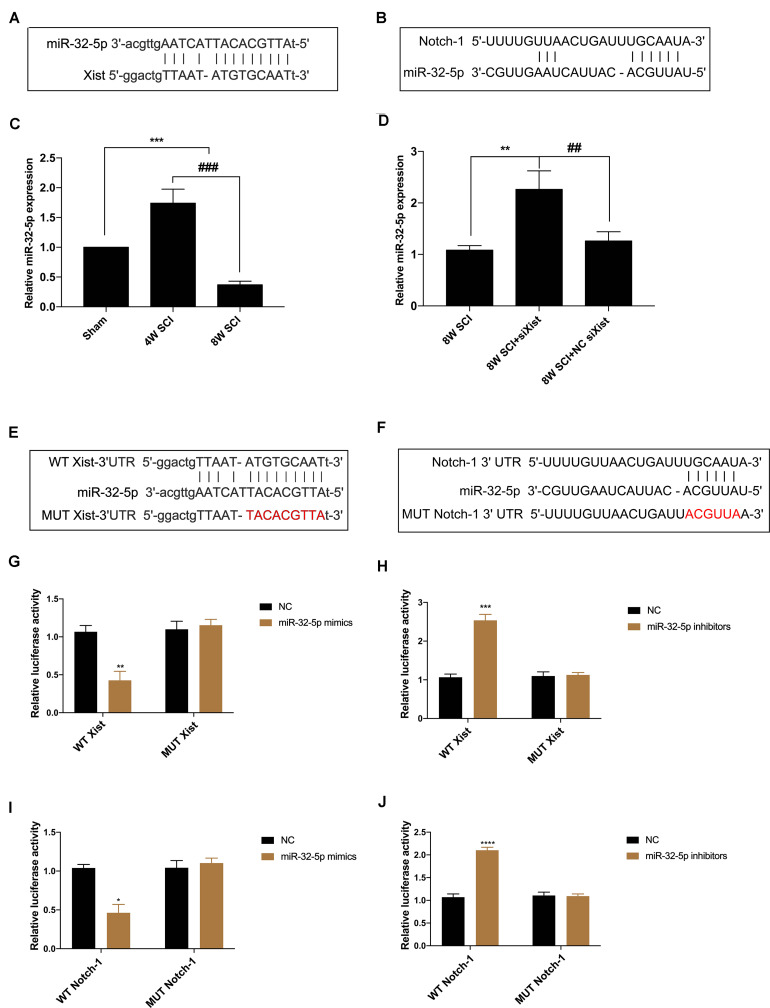
FIGURE 4.
Xist and Notch-1 are direct targets of miR-32-5p. (A) Binding regions between miR-32-5p and Xist. Bioinformatics analysis (ChipBase, LncRNAdb, and StarBase) was used to predict the target microRNA regulated by Xist. (B) Binding regions between Notch-1 and miR-32-5p. Bioinformatics analysis (TargetScan, Starbase, miRanda, and miRDB) was used to predict the putative target of miR-32-5p. (C) Quantitation of the relative miR-32-5p expression in the sham, 4W SCI, and 8W SCI groups by RT-qPCR. (D) Quantitation of the relative miR-32-5p expression in the 8W SCI, 8W SCI + siXist, and 8W SCI + NC siXist groups by RT-qPCR. (E) Dual-luciferase reporter constructs containing the wild-type (WT Xist) or mutant (MUT Xist) Xist sequence. (F) Dual-luciferase reporter constructs containing the wild-type (WT Notch-1) or mutant (MUT Notch-1) Notch-1 sequence. (G) Dual-luciferase reporter assay of miR-32-5p and Xist following transfection of miR-32-5p mimics. HEK-293T cells were co-transfected with the miR-32-5p mimics or corresponding NC and the dual-luciferase vector containing WT Xist or MUT Xist and incubated for 48 h. (H) Dual-luciferase reporter assay of miR-32-5p and Xist following transfection with miR-32-5p inhibitors. HEK-293T cells were co-transfected with the miR-32-5p inhibitors or the corresponding NC and the dual-luciferase vector containing WT Xist or MUT Xist. (I) Dual-luciferase reporter assay of Notch-1 and miR-32-5p following transfection of miR-32-5p mimics. HEK-293T cells were co-transfected with the miR-32-5p mimics or the corresponding NC and the dual-luciferase vector containing WT Notch-1 or MUT Notch-1 and incubated for 48 h. (J) Dual-luciferase reporter assay of miR-32-5p and Xist following transfection with miR-32-5p inhibitors. HEK-293T cells were co-transfected with miR-32-5p inhibitors or the corresponding NC and the dual-luciferase vector containing WT Xist or MUT Xist. Three independent experiments were carried out. *p < 0.05, **p < 0.01, ***p < 0.001. ****p < 0.0001; ##p < 0.01, ###p < 0.001. Data are expressed as the mean ± SEM.