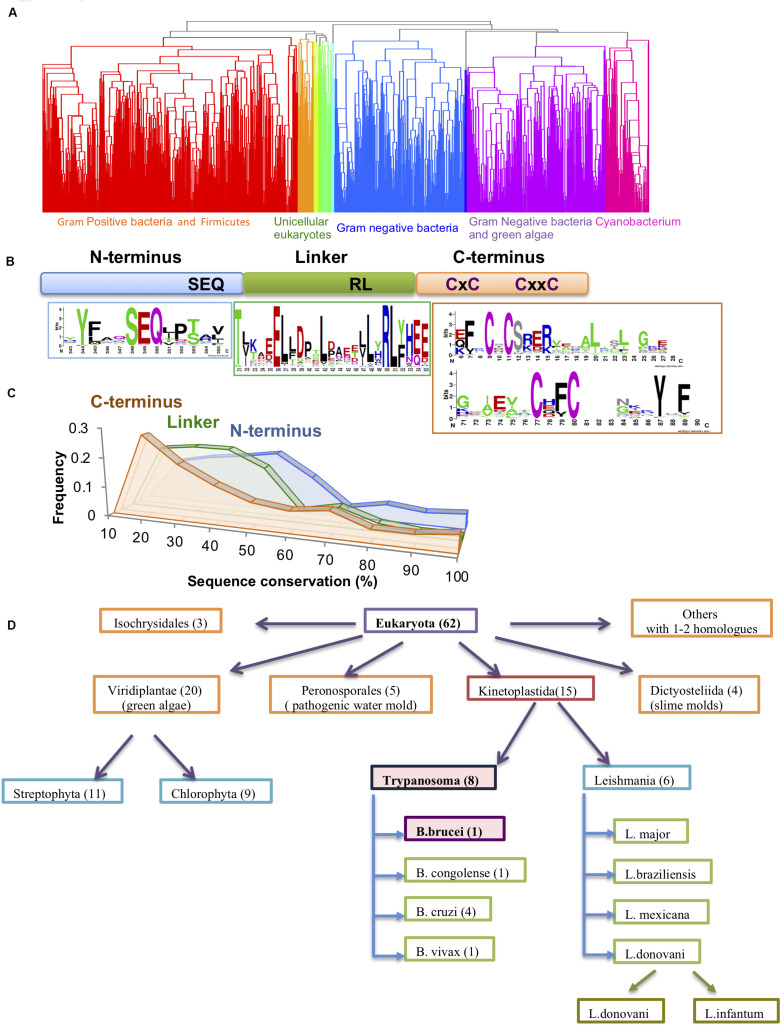
FIGURE 1.
Conservation of the Hsp33 protein family. (A) Clustering of the 1579 Hsp33 homologs according to their sequence conservation derived by hierarchical clustering as described in the method part. Ten obtained clusters (shown in different colors) correspond to the phylogenetic origin of the sequences. (B) Schematic representation of the Hsp33 domains. Sequence motifs of the most conserved sequence regions are shown in sequence logos created by WebLogo (Crooks et al., 2004). (C) Comparison of sequence conservation between the three Hsp33 regions [N-terminus (blue), linker (green) and C-terminal redox domain (orange)]. The plot shows the distribution of the position-based amino acid conservation scores as described in section “Materials and Methods.” (D) Hsp33 orthologs in eukaryotes. Kinetoplastida and Viridiplantae families contain the larger numbers of Hsp33 orthologs.