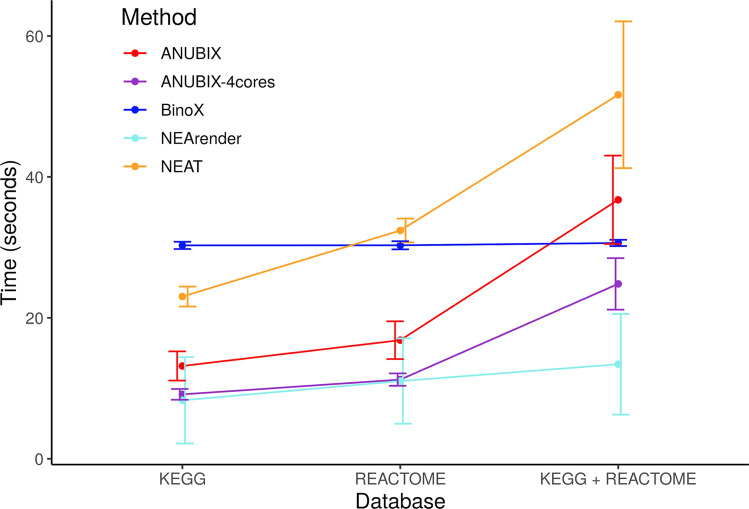
Figure 8.
Compute time when running a random experimental gene set from MSigDB. 100 different gene sets were tested against KEGG, REACTOME and KEGG plus REACTOME pathways, for each of the network-based methods. Since ANUBIX allows parallelization we also added another run with 4 cores. The error bars show the variability in compute time for each of the methods in each of the databases. The BinoX precomputation step is not included since it takes 22 min. Software: R version 3.4.3 https://www.r-project.org/.