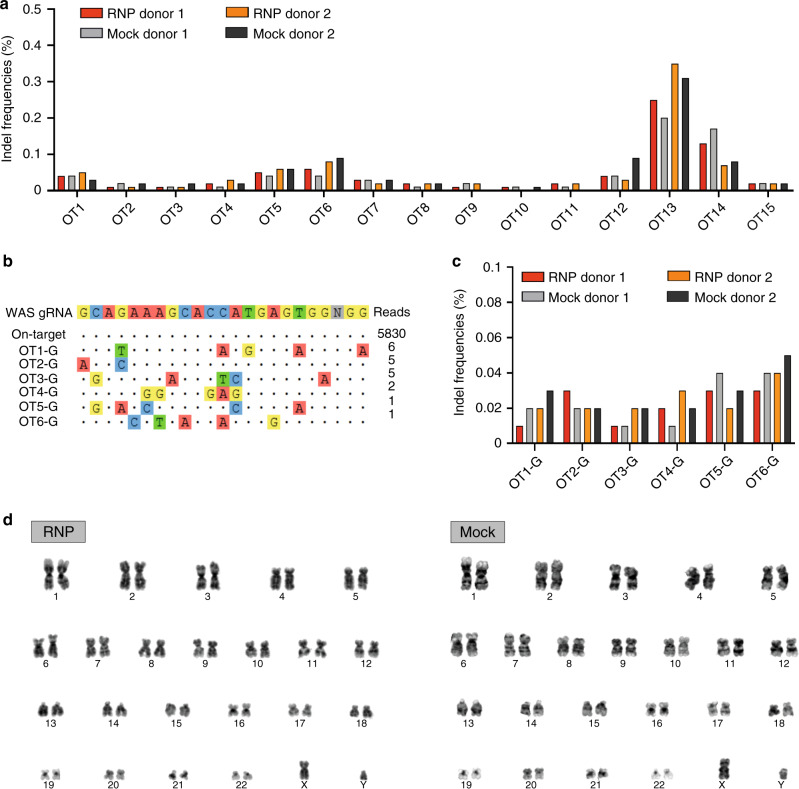
Fig. 6. Genotoxicity analysis in edited HSPCs.
a Targeted high-throughput sequencing of off-target sites in edited (RNP) or electroporated-only (Mock) HSPCs (n = 2 experiments from two different donors; no significant difference between RNP and Mock samples when pooled and analysed with two-tailed paired Student’s t test). b Sequences of off-target sites identified by GUIDE-seq for WAS gRNA. The intended target sequence is shown in the top line with mismatches to the on-target site shown and highlighted in colour. GUIDE-seq sequencing read counts are shown to the right of each site. GUIDE-seq sequencing read counts are shown to the right of each site. c Targeted high-throughput sequencing of off-target sites in edited (RNP) or electroporated-only (Mock) HSPCs (n = 2 experiments from two different donors; no significant difference between RNP and Mock samples when pooled and analysed with two-tailed paired Student’s t test). d Karyotype analysis of unmanipulated (Mock) and genome edited (RNP) HSPCs showed no chromosomal abnormalities (100 cells analysed for each condition). Source data are provided as a Source Data file.