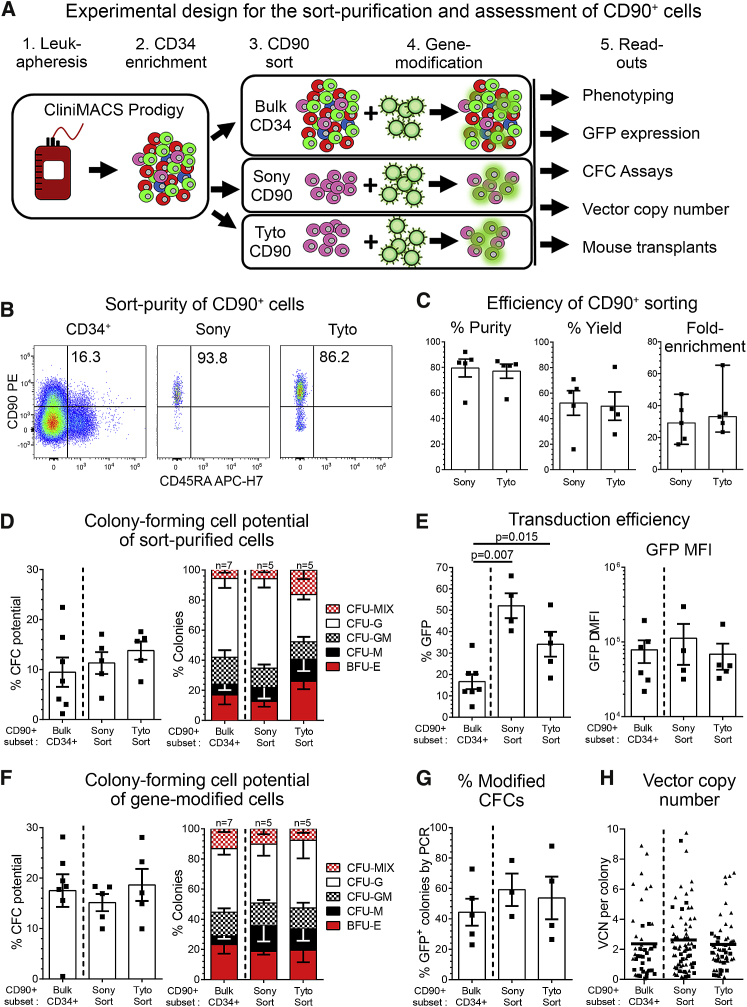
Figure 5.
FACS Purification and Quality Control of CD90+ HSPCs
(A) Schematic of the experimental design. (B) Flow cytometric assessment of cells before FACS (CD34+, first plot) and purified CD90+ cells after sorting on the Sony (second plot) and Tyto (plot). (C) Comparison of the purity, yield, and fold enrichment of CD90+ HSPCs on the Sony and Tyto sorters. (D) CFC potential of CD90+ cells within bulk CD34+ HSPCs and FACS-purified CD90+ subsets. CD90+ cells from all three conditions were sorted into CFC assays to exclude contaminating CD90– cells. (E) Flow cytometric quantification of GFP-expressing CD90+ cells within bulk CD34+ cell and FACS-purified CD90+ subsets (left) and the delta-MFI of GFP expression in gene-modified cells (right). (F) Erythroid, myeloid, and erythro-myeloid CFC potential of gene-modified CD90+ cells from bulk CD34+ cells and FACS-purified CD90+ subsets. CD90+ cells from all three conditions were sorted into CFC assays. (G and H) Individual colonies from all three conditions in (F) were picked, and (G) the gene-modification efficiency in CFCs was determined by PCR as well as (H) the VCN in modified CFCs quantified by qPCR. (See also Table S9.) Statistics, means ± SEM; in C, third graph, median and range; significance values, two-tailed paired t test.