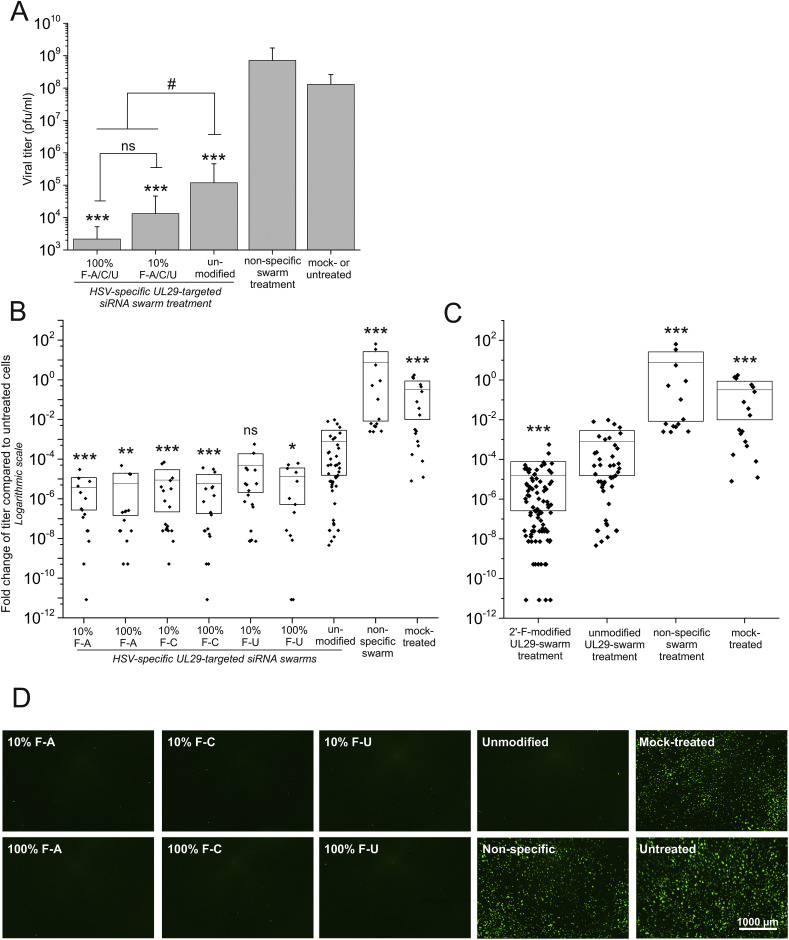
Fig. 3.
Antiviral activity of siRNA swarms is improved by 2′-F-dNMP modifications. U373MG cells were infected with 1000 pfu of HSV-1-GFP (MOI≈0.06) at 4 h after treatment with each of the UL29-2′-F-siRNA swarms (50 nM), unmodified UL29-siRNA swarm (50 nM) or non-specific swarm (50 nM). The guide strand of modified siRNA swarms was fully (100%) or partially (10%) modified and contained 2′-F-dAMP, 2′-F-dCMP or 2′-F-dUMP as the modified nucleotide (10% or 100% F-A, F-C or F-U, respectively). The controls were transfected with water only (mock-treated) or were left untreated. At 48 hpt, the culture supernatants were collected for quantitation of the virus (A, B, and C), and live cell imaging was performed (D). (A) Viral titer (pfu/ml) was determined from the supernatant samples by plaque titration. The samples treated with partially modified (10%) 2′-F-siRNA swarms (A, C, or U), and those treated with fully (100%) modified 2′-F-siRNA swarms were grouped for statistical analysis. The error bars represent standard deviation of the mean from two separate experiments with at least three replicate samples of all individual treatments and controls (N = 15–33 or N = 6–12, respectively). The significance of the changes is shown against both the non-specific swarm as well as the mock- and untreated samples (***, p ≤ 0.001). The significant changes against a group of comparison are marked with # (#: p ≤ 0.05, ns: non-significant). (B, C) Viral shedding quantitated from the culture supernatants is shown for each individual siRNA swarm (B) or collectively for the modified siRNA swarms (C). The fold change of viral titer is presented in comparison with the untreated samples. The efficacies of the siRNA swarms, with the indicated modified nucleotides, are presented separately (N = 13–18) in panel B and collectively (N = 96) in panel C. The mean and standard deviation (shown with a horizontal line and a box, respectively) for each sample group represent five separate experiments (N = 13–96). The diamonds illustrate the individual samples. Significant changes against the unmodified UL29-specific swarm are marked with asterisks (*: p ≤ 0.05, **: p ≤ 0.01, ***: p ≤ 0.001, ns: non-significant). (D) Representative fluorescent images of the HSV-1-GFP -infected cells with the indicated siRNA swarm or control treatments were taken at 48 hpt. The scale bar represents 1000 μm.