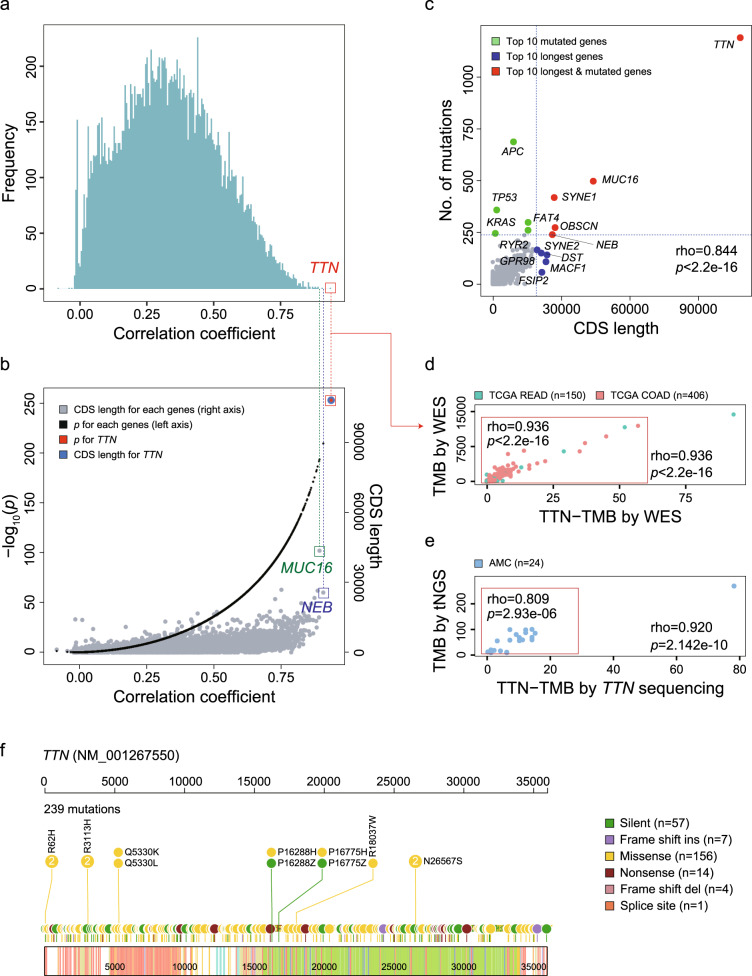
Fig. 2. Correlation analysis of two independent colorectal cancer cohorts.
a Histogram of the correlation coefficients between the mutation count of each gene and the total mutation count in a TCGA colorectal cancer (CRC) cohort (n = 556) (Pearson correlation test). b p-values (black dots) for the correlation coefficient and CDS lengths (gray dots) for each gene (Pearson correlation test). c Comparison of CDS lengths and mutation counts in all observed genes within the TCGA CRC cohort. Red dots indicate top genes possessing both high CDS lengths and frequent mutation, blue dots indicate genes with relatively large CDS lengths and infrequent mutation, and green dots indicate genes with relatively short CDS lengths and frequent mutation. Correlation of TTN-TMB, TMB-WES, and TMB-tNGS in the TCGA CRC cohort (d) and the AMC cohort (e) (Pearson correlation test). f Lollipop plot of all reported variants of the TTN gene in the AMC cohort. TMB tumor mutation burden, WES whole-exome sequencing, tNGS targeted next-generation sequencing.