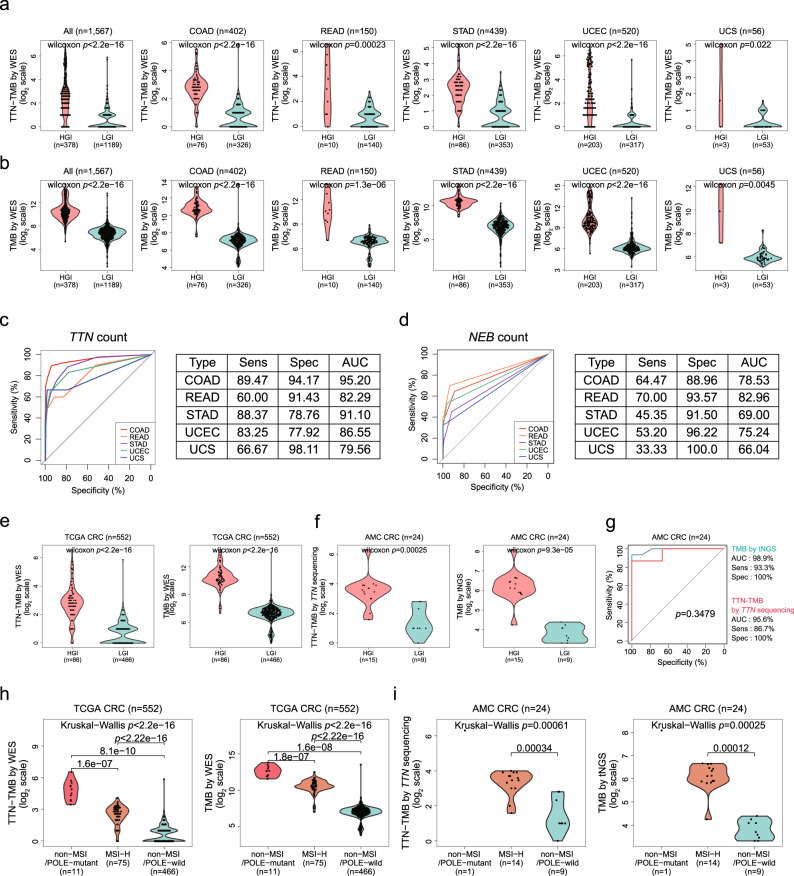
Fig. 5. Comparison of TTN-TMB according to MSI and POLE mutation status.
Mutation counts within the TTN gene (a) and total mutation counts of all genes (b) according to HGI and LGI samples in the TCGA cohort (n = 1567, Wilcoxon rank-sum test) for each tumor type. Diagnostic performance of TTN (c) and NEB (d) for HGI in each tumor type. The differences in TTN mutation count between the HGI and the LGI groups in the TCGA CRC cohort (e) and the AMC cohort (f) (Wilcoxon rank-sum test). g No differences in diagnostic accuracy for HGI and LGI were observed between TTN-TMB and TMB by targeted NGS (tNGS) in the AMC cohort. The differences in TTN mutation count among the POLE-mutated group, the MSI-H group, and the remaining group in the TCGA CRC cohort (h) and the AMC cohort (i) (Wilcoxon rank-sum test or Kruskal-Wallis test). HGI high genomic instability, LGI low genomic instability, TMB tumor mutation burden, WES whole-exome sequencing, NGS next-generation sequencing, AUC area under the curve.