Abstract
Glass nanopipette has shown promise in applications for single-cell manipulation, analysis, and imaging. In recent years, plasmonic nanopipette has been developed for single-cell analysis to take the advantage of surface-enhanced Raman spectroscopy (SERS) measurement. In this work, we developed a SERS-active nanopipette that can perform long-term and reliable intracellular analysis of single living cell with minimal damage, which is achieved by optimizing the nanopipette geometry and the surface density of gold nanoparticle (AuNP) layer at the nanopipette tip. To demonstrate its capability in single-cell analysis, we have applied the nanopipette for intracellular pH sensing. Intracellular pH (pHi) is vital to cells as it influences cell functions, behaviors and pathological conditions. The pH-sensitivity was realized by simply modifying the AuNP layer with pH reporter molecule 4-Mercaptobenzoic acid. With a response time of less than 5 seconds, the pH sensing range is from 6.0 to 8.0 and the maximum sensitivity is 0.2 pH units. We have monitored the pHi change of individual HeLa and Fibroblast cells, triggered by the extracellular pH (pHe) change. The HeLa cancer cell can better resist pHe change and adapt to the weak acidic environment. The plasmonic nanopipette can be further developed to monitor other intracellular biomarkers.
Graphical Abstract
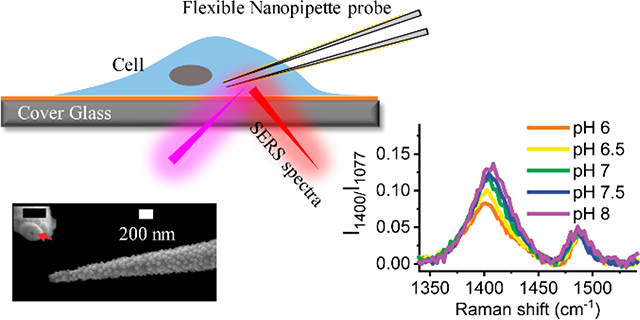
SERS-active flexible nanopipette can conduct long-term reliable intracellular single-cell analysis
Introduction
Reliable detection and quantitative analysis of biomarkers at a single cell level are critical and vital for detecting diseases earlier and understanding the fundamental biological process better. Yet, a low concentration of biomarkers, micron dimension (around 1–50 μm), and the dynamic nature of living cells make single-cell intracellular content analysis challenging with traditional analytical methods.1, 2 In the past several decades, several analytical techniques have emerged to overcome the challenges of single-cell intracellular sensing, including fluorescence-based spectroscopy3/microscopy4, dark-field scattering microscopy,5 surface-enhanced Raman spectroscopy (SERS),6 and nanopore/nanoelectrode sensing.7 Fluorescence-based spectroscopy/microscopy techniques have been routinely performed for single-cell analysis as they are capable of tracking intracellular molecules individually with high spatial and temporal resolution. However, only limited number of cell-permeant fluorophores8 can be selected or gene construction technique9 is required to allow intracellular sensing of live cells. Also, the fluorophore has intrinsic stability problem associated with photobleaching and blinking over long time scales.10–12 SERS is an attractive alternative to fluorescence-based spectroscopy/microscopy techniques for biological sensing and has gained increasing attentions.13 SERS signal is more photo-stable and provides multiple narrow bands at the same time, which allowing unparalleled multiplex detection capability. In the common biological sensing of SERS technique,14, 15 untethered plasmonic nanoparticles (NPs) enter the cell through endocytosis to enable intracellular SERS detection. The advantage of untethered method is that it is convenient to use and less invasive to cells. However, the cellular retention time of NPs is typically long, and the intracellular distribution of these nanoprobes is uncontrollable.
To address the problems associated with the uncontrollable delivery of untethered NPs, a tethered substrate was used for “point-and-shoot” single-cell analysis. The tethered substrates include carbon nanotube,16, 17 fiber-optic tip,18 nanopipette, nanoelectrode,19 and nanopore.20, 21 Glass micropipette has been widely used in single ion-channel measurement in electrophysiology. In recent years, nanopipette with the apex size of a few hundred nanometers or less have been applied for intercellular measurements. Pourmand’s group has pioneered the nanopipette robotic ‘nanobiopsy’ system for single-cell genome sequencing.22 The nanopipette is also compatible with the traditional patch-clamp system. Flexible nanopipettes have been used to perform in vivo electrophysiology in living cell or tissue with minimized cell damage.23, 24 Because of the advantages of SERS measurement, plasmonic nanopipettes have also been fabricated to enable SERS based intracellular measurements.25–30 Although promising, the development of plasmonic nanopipettes for intracellular applications is still at the early stage. For example, to generate enough SERS signal for sensing, the tip size of the plasmonic nanopipette is generally bigger, which may cause bigger cellular damage during the insertion of plasmonic nanopipette tip. Therefore, to enable long-term time-resolved intracellular measurement, it is important to optimize both the nanopipette geometry and the plasmonic substrate structure to the balance the needs of enhancing SERS signal and minimizing cell damage.
Herein, we developed a SERS-active nanopipette for intracellular pH (pHi) sensing for individual eukaryotic cells. The pHi is an important parameter for regulating cellular functions, behaviours and pathological conditions.31 However, effective intracellular sensors are still limited.32 To minimize the cellular damage during the insertion of the nanopipette, we reduced the naonopipette apex size to below 200 nm. The geometry of the nanopipette has also been optimized to increase its flexibility to better match the stiffness of the cell. By optimizing the distribution, density, and surface chemistry of the adsorbed AuNPs on the outer surface of the nanopipette, we obtain a uniform, stable, and reproducible SERS-active substrate with a high enhancement of Raman intensity. Molecule 4-mercaptobenzoic acid (4-MBA) was used as the probe molecule and the ratiometric intensity signal of COO- stretching vibration mode was calibrated for pH sensing. To evaluate the dynamic response of the nanopipette to pH changes in real-time, the nanopipette was first tested in the controlled fluidic flow in a microfluidic device. Then, the nanopipette was applied to study the pHi change of individual live cells. By changing the extracellular pH (pHe), we compared the cytoplasmic pH changes between cancer cells and normal cells.
Experimental
Reagents and materials
4-Mercaptobenzoic acid (4-MBA, 99 %) and (3-Aminopropyl) triethoxysilane (APTES, >98 %), sodium chloride (NaCl, >99 %), potassium phosphate dibasic (K₂HPO4), potassium phosphate monobasic (KH₂PO₄) were purchased from Sigma-Aldrich. Phosphate buffered saline (PBS) powder (for pH 7.3−7.5), potassium chloride, absolute ethanol (200 proof), and reagent alcohol (histology grade), hydrochloric acid (HCl, ~37 %) and Sodium hydroxide (NaOH) were purchased from Fisher Scientific. Citrate protected 40 nm gold nanoparticles (AuNPs) colloid was purchased from Ted Pella, Inc.. All the aqueous solutions were prepared using deionized (DI) water (~18 M ohm, Ultra Purelab system, ELGA/Siemens).
The batch solution pH ranging from 4.3 to 10.1 for nanopipette nanoprobe calibration was obtained by adding HCl or NaOH. The pH ranging from 6.0 to 8.0 was obtained by changing the volume ratio of the K2HPO4 and KH2PO4, as shown in table ESI9, ESI†. The pH values were measured by using a pH meter (SympHony SR60IC, VWR International).
Fabrication of pH-sensitive, SERS-active, and flexible nanopipette
The borosilicate glass capillary with filament (O.D.: 1.0 mm, I.D.: 0.58 mm, 15 cm length, Sutter Instrument Co.) was cleaned by Piranha (caution: Piranha solutions are highly corrosive and need to be handled with extreme caution!) for 30 min. Then rinsed by DI water both inside and outside thoroughly and dried in an oven at 120 °C for overnight. Glass long-taper (flexible) and short-taper (stiff) nanopipettes were prepared from the cleaned glass capillaries by using a laser-based pipette puller (P-2000, Sutter Instrument) with different parameters. Parameters for long-taper: HEAT = 500, FIL = 4, VEL = 50, DEL = 255, PUL = 100. Parameters for short-taper: HEAT = 400, FIL = 4, VEL = 50, DEL = 255, PUL = 150.
The prepared glass nanopipette was soaked in 0.4 % (v/v) ethanol solution of (3-Aminopropyl) triethoxysilane (APTES) for 1.5 hrs. After rinsing with ethanol, the nanopipettes were dried by the argon gas flow. Subsequently, APTES modified nanopipette was immersed in 150 pM AuNPs with 40 nm diameter for a different time at 4 °C in the refrigerator. At last, the AuNP-loaded glass nanopipette was soaked in ethanol solution with 1 mM 4-MBA for 1 hr. After cleaning, the prepared nanopipette was stored in 1x PBS before use.
Cell culture
HeLa cells and fibroblasts from mouse cells were cultured in Dulbecco’s Modified Eagle Medium (Gibco) with 10 % fetal bovine serum at 37 °C, 5 % CO2 and 90 % humidity. The cells were seeded on the cover slides (pre-modified with 0.01 % poly-L-lysine to increase the cell adhesion). Before SERS measurement, the cells were gently washed two times with 1xPBS and the culture medium were supplied with K2HPO4/KH₂PO₄ buffer with different pH values. 4 % paraformaldehyde was used to fixed live cells for 10 min and rinsed with 1xPBS.
SERS experiments
Micromanipulator (Thorlabs, PCS-5300) was used to control the position of the SERS-active nanopipette tip for single-cell measurements. SERS was performed on a home-built Raman microscopy setup (see details in reference33) using a Nikon inverted optical microscope. The 632.8 nm laser beam was focused on the nanopipette apex that was immersed in a liquid cell on the microscope sample stage. Neutral-density filters attenuated the laser power to 1 mW and the laser spot size was about 3 μm. Thus, the typical area intensity was about 35 μW μm−2. Time-resolved SERS spectra were collected with an acquisition time of 1–2 seconds per frame and accumulated 30 times for each spectrum. The spectral resolution was about 2 cm−1.
Results & Discussion
Fabrication and characterization of the flexible nanopipette-based nanoprobe
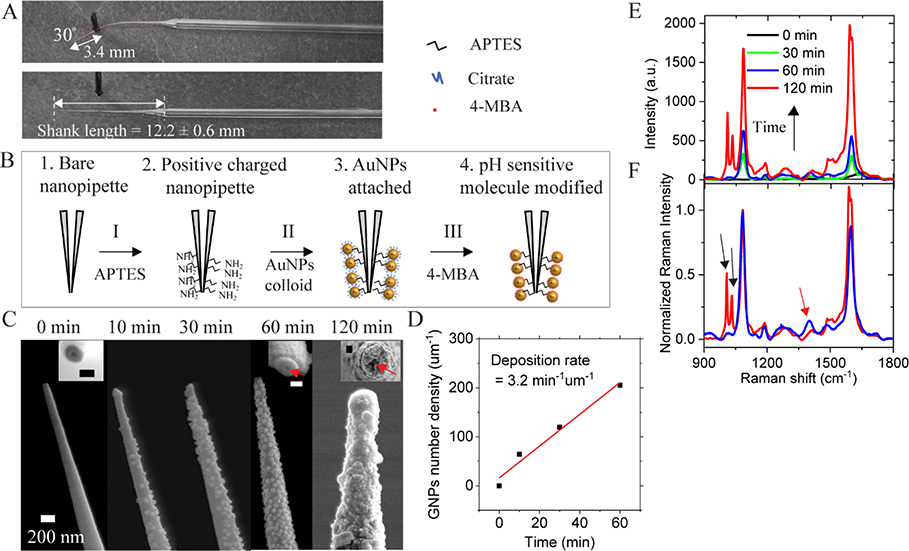
Nanopipette has been successfully used in intracellular studies. However, the insertion of nanopipette into small mammalian cells may damage the cell membrane.34, 35 With the aim of minimizing the cellular damage, both the apex size and the stiffness of the nanopipette tip are optimized. In a recent report, the nanopipette showing minimized invasive in the intracellular measurement has a spring constant of 0.08 N/m.23 Therefore, we prepared nanopipettes with a long-taper (shank length ~12.2 mm) geometry and a small tip diameter (sub-200 nm) using a pipette puller. Based on the classic beam theory, the spring constant of the nanopipette tip is estimated about 0.056 N/m with the cantilever approximation (See details in ESI S1, ESI†). In order to demonstrate the flexibility of the long-taper nanopipette, the nanopipette tip is bent ~ 30° at a distance ~3.4 mm from the apex and is then fully recovered after relaxing, as shown in Fig. 1A. The flexible tip can be repeatedly bent without breakage. In addition, the insertion of the flexible nanopipette tip into the living cell only induces minimal damages, which will be discussed in the single-cell intracellular pH sensing section. It demonstrates that the flexible long-taper nanopipette is preferable for intracellular measurements.
Fig. 1.
(A) Optical images demonstrate the flexibility of a long-taper nanopipette tip. (B) Scheme of the procedures for surface modification of a pH-sensitive nanopipette (not to scale). (C) SEM images of the nanopipette tip with different AuNP deposition time, ranging from 0 to 120 min. The inset images are the top view of the nanopipette orifice. The scale bar is 100 nm in inset images. (D) The AuNP number density at the tip (2 μm from the apex) as a function of the AuNP deposition time. (E) Typical Raman spectra of 4-MBA from the AuNP-loaded nanopipette tip after different AuNP deposition time. (F) Normalized Raman spectra of 4-MBA after 60 min (blue line) and 120 min (red line) AuNP deposition time.
After the fabrication, we modified the nanopipette tip to be a SERS-active substrate. The schematic diagram in Fig. 1B shows the steps to prepare a pH-sensitive and SERS-active nanopipette. The details are given in the Methods section. Briefly, the tip of the nanopipette was first modified with APTES. Then the nanopipette was soaked in an aqueous solution containing AuNPs for various deposition time. Consequently, the outer surface of the nanopipette was coated with a layer of AuNPs through electrostatic force. Lastly, 4-MBA molecules were modified to the AuNPs.
The previous simulation results36 have shown that the optimization of the SERS substrate can be achieved by controlling the AuNP coverage. We altered the AuNP deposition time to control the surface coverage of the AuNPs on the nanopipette tip while keeping the conditions of other steps the same. Fig. 1C shows the SEM images of the nanopipette tip after immersing the APTES modified nanopipette in colloidal AuNP solution for different time intervals. Before the deposition of AuNPs, the outer and inner diameters of the nanopipette are around 100 nm and 30 nm, respectively. After 60 min of deposition, the outer diameter of the nanopipette is increased to approximately 170 nm, revealing the adsorption of one sublayer of 40 nm AuNP. The inner channel remains open, as indicated by the red arrow in Fig. 1C. After 120 min of deposition, the AuNP layer becomes much thicker with a multi-layer configuration. The inner diameter of the pore is also significantly reduced or even entirely blocked. Based on the SEM images, we can estimate the average number density of AuNPs on the nanopipette tip. As shown in Fig. 1D, the surface number density of AuNP increases linearly at a rate of 3.2 min−1 μm−1 during the deposition of the first 60 min. At 60 min, the surface number density of AuNP is about 202 μm-2.
After modifying 4-MBA molecules to the AuNP surface, Raman spectra were taken at the tip of the AuNP-loaded nanopipette in 1xPBS. As shown in Fig. 1E, the overall 4-MBA Raman intensity increases with the increase of the number density of AuNP at the tip. The nanopipette with a higher AuNP density possesses more plasmonic AuNPs and nanogaps, inducing a higher electromagnetic enhancement of SERS intensity. Fig. 1F shows the same Raman spectra of 4-MBA in Fig. 1E normalized to the peak at 1077 cm−1. The band at 1400 cm−1 Raman spectra (indicated by the red arrow) is used for pH sensing. This band will be further discussed in the next section. When the AuNP number density is too high (the red color spectrum), the intensity of this band decreases obviously along with the appearance of two new bands37, 38 near the 1077 cm−1 peak (indicated by the black arrows). Thus, there is an optimal thickness for the AuNP layer. We have determined that the thickness of AuNP layer after 60 min of AuNP deposition is the best for pH sensing.
The pH calibration of the flexible nanopipette-based nanoprobe
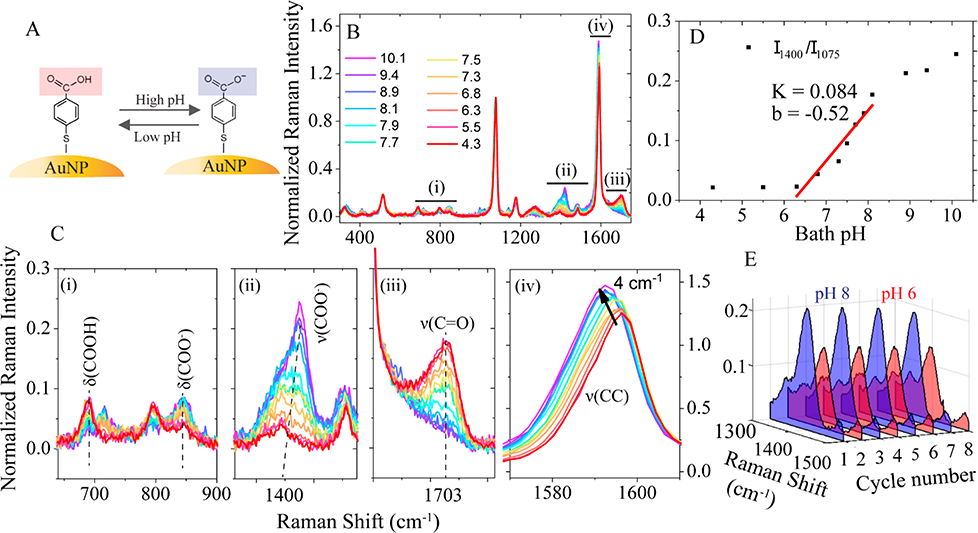
Fig. 2A illustrates the pH-sensing mechanism of 4-MBA molecule. The carboxyl group of 4-MBA is protonated at a lower pH and deprotonated at a higher pH, leading to changes in the Raman spectrum. Fig. 2B shows the pH-dependent Raman spectra of 4-MBA, collected from a fabricated pH-sensitive nanopipette. To avoid sample-to-sample variations, we used the ratiometric method to characterize the pH-dependent response of the SERS signal. Here, the band at 1077 cm−1, from the stretching mode of C-S coupled with the benzene ring, is used as the internal reference. All other bands in the SERS spectra were normalized to the internal reference (all SERS spectra shows after this point were normalized if not mentioned otherwise). The bath solution pH value varies from 4.3 to 10.1 results in the systematic changes in four major spectral regions, as marked with numbers (i) to (iv) in Fig. 2B.
Fig. 2.
(A) The schematic of reversible changes of the carboxyl group of 4-MBA as a result of pH changes. (B) The SERS spectra of the nanopipette-based nanoprobe in bath solution with different pH values. (C) ((i)-(iv)) The zoom-in of four pH-sensitive spectral regions marked in (B). (D) pH calibration curve obtained by plotting the ratio of 1400 cm−1 against different pH values. The black dots are fitted by linear regression equation and plotted as a solid red line. (E) The 3D SERS spectra of the normalized intensity change of band 1400 cm−1 when the bath solution pH was alternated between 8.0 and 6.0 for 8 cycles.
Fig. 2C shows the zoom-in of the four pH-sensitive spectral regions. We assigned the Raman bands based on the literatures39–42 and our density functional theory (DFT) calculation (ESI S2, ESI†). Fig. 2C(i) shows the bands at 688 and 847 cm−1, which are attributed to the bending mode of δ(COOH) and δ(COO-). The two peaks decreased and increased concurrently with pH due to the deprotonation of -COOH, respectively. However, these bands were often overlapped with the bands from the glass substrate (see details in Fig. S3, ESI†). Fig. 2C(ii) shows the pH dependent change of band at 1400 cm−1, which is assigned to the symmetric COO- stretching mode (ν(COO-)). Its intensity increases with the increase of solution pH. Figure 2C(iii) shows the pH-dependent band at 1703 cm−1, which is attributed to the stretching mode of the carbonyl group (ν(C=O)). Its intensity decreases with the increase of solution pH. However, if the overall SERS intensity is weak, like the case of intracellular measurement, the bending mode of water molecules at 1640 cm−1 may interfere with this mode (see Fig. S3, ESI†). Fig. 2C(iv) shows the change of band at 1592 cm−1. This band redshifts (to the lower wavenumber) 4 cm−1 as the solution pH increases from 4.3 to 10.1, which is due to the breaking of symmetry upon the deprotonation of the carboxyl group[30]. However, this Raman shift is too small considering the 2 cm−1 spectral resolution of our setup. However, this band can be useful if we increase the spectral resolution of the spectrometer.
All four regions can be used to detect local pH changes for different applications. Among them, we find the band at 1400 cm−1 in Fig. 2C(ii) is the most robust and sensitive peak for the cellular application. Therefore, we use this band for the following experiments. Fig. 2D shows the plot of intensity ratio I1400/I1077 versus bath solution pH, in which the most sensitive pH range is from 6.0 to 8.0. A linear fit to the data in this dynamic range gives a slope of 0.084. The noise level of the normalized intensity is about 0.015, which gives the pH sensitivity of 0.2 units. The measured slope values vary from 0.029 to 0.084 for different nanopipettes. Therefore, it is important to obtain the calibration curve for each nanopipette. The reproducibility of the nanopipette for pH sensing was also tested. Fig. 2E demonstrates the spectral changes of the normalized intensity of the v(COO-) mode between pH 6.0 (red color) and 8.0 (blue color) is reproducible.
Dynamic response of the nanopipette-based pH nanoprobe to pH changes
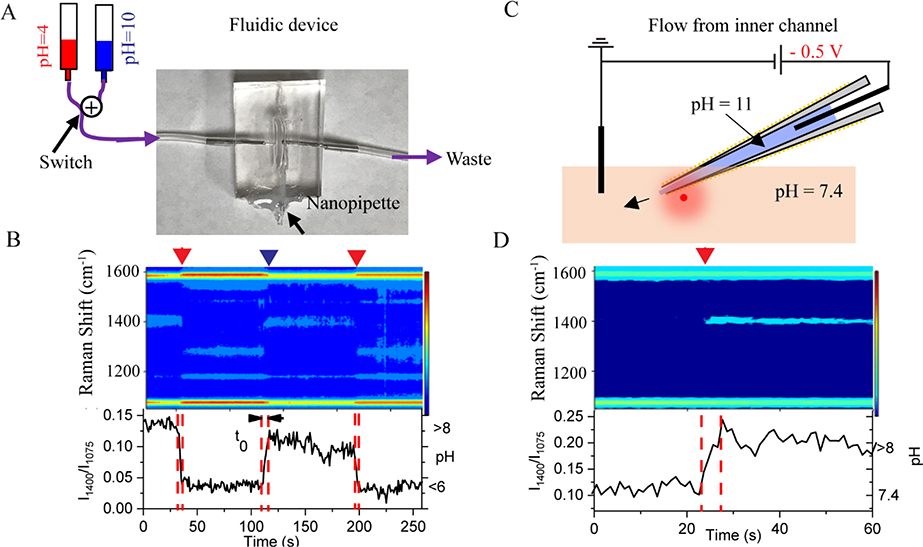
Motivated by the temporal dynamics of living cells, it is essential to understand the response time of the nanoprobe for dynamic pH changes in the solution. Fig. 3A shows the fluidic device made by Polydimethylsiloxane (PDMS). The nanopipette is embedded in the middle of the fluidic chamber. By switching two solutions with pH 4 and 8 at the inlet, controllable pH changes are introduced in the fluidic chamber. The measured real-time SERS changes are shown in the heatmap trajectory in Fig. 3B. From the intensity (I1400/I1077)-time trace at the bottom panel of Fig. 3B, it takes about 5 sesconds (indicated as t0 in Fig. 3B) to reach the maximum signal in responding to the solution pH change. This response time is mainly limited by the fluid flow speed. It takes about 5 s for the solution to fully fill the 1000 μL fluidic chamber with a speed of 200 μL/s while the time resolution of the Raman signal is 1 s. Nevertheless, the results imply that the nanopipette-based pH nanoprobe has a pH response time at least of a few seconds.
Fig. 3.
(A) Schematic (not to scale) of a fluidic device with the embedded nanopipette-based nanoprobe. The bottom of the fluidic device is sealed by a cover glass. Solution with two different pH values passes through the nanopipette alternatively. (B) Top: Dynamic responding of SERS heatmap as two different pH bulk solution passing through nanopipette. Red and blue arrows indicate the moment that pH 4.0 and pH 10.0 bulk solution passing to nanopipette. Bottom: intensity ratio (I1400/I1077) time trace of and right y-axis indicate pH value. (C) Schematic drawing (not to scale) of pH 11 solution filled nanopipette exchange H+ with pH 7.4 surrounding solution. (D) Top: Dynamic responding of SERS heatmap as H+ exchange through nanopipette open channel. Red arrow indicates the moment of −500 mV potential applied. Bottom: intensity ratio (I1400/I1077) time trace and right y-axis indicate pH by using the calibration curve. The Raman spectra were acquired at 1 s/frame.
The open channel of the nanopipette can also be utilized for pH sensing. A proof-of-concept experiment is illustrated in Fig. 3C. The pH of the solution inside the nanopipette barrel is 11, and the pH of the bath solution is 7.4. By applying a negative bias at the Ag/AgCl electrode inside the nanopipette barrel, the hydroxyl ions inside the nanopipette are driven out of the nanopieptte barrel by the electric force. The effect of bias driven local pH change is demonstrated in the SERS trajectory shown in Fig. 3D. At zero bias, the SERS trajectory was very stable and no spectral changes are detected (See Fig. S4, ESI†). After applying −0.5 V, the peak near 1400 cm−1 increased significantly, suggesting the increase of local pH. Therefore, the concentration gradient alone is not enough to drive hydroxyl ions out of the nanopipette tip. The applied bias is effective to deliver hydroxyl ions from the nanopipette tip to the bath solution. This experiment demonstrates the SERS active nanopipette loaded with molecules/ions inside the barrel could be used to electrically regulate the molecule/ion delivery while simultaneously monitoring the real-time and in-situ changes induced by the delivered molecules/ions through Raman spectral changes.
Single-cell intracellular pH Sensing
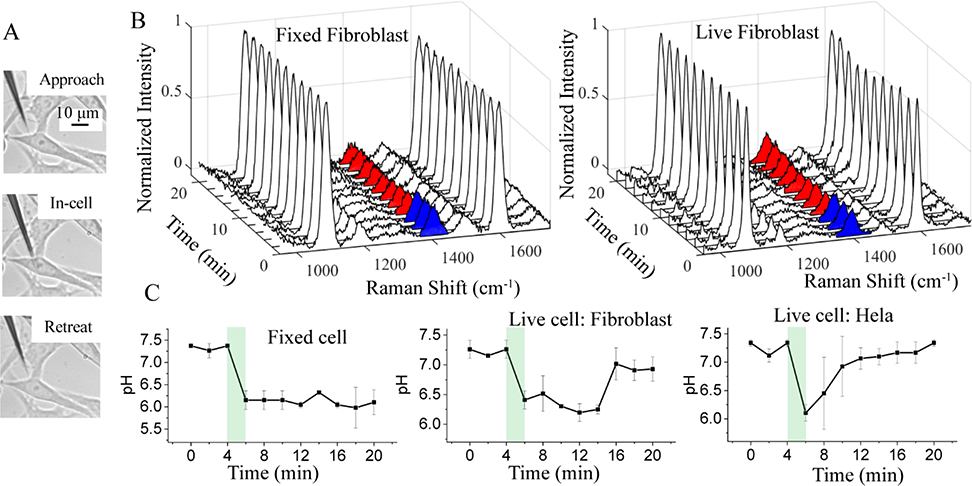
We further use the pH-sensitive nanopipette to detect pHi changes in individual cells. Controlled by a piezo micromanipulator, the pH-sensitive nanopipette tip is inserted into HeLa and Fibroblast cells at the center of the cells. Time-resolved SERS measurements are performed when the nanopipette tip reaches the designed location. Fig. 4A shows the optical images of HeLa cells when the nanopipette tip was approached to, inside, and retreated out of the cell, respectively. The similar optical microscope images for the inerstion process of nanopipette at the fibroblast cell are shown in Fig. S5, ESI†. The cell damage induced by the insertion of the nanopipette was tested by the Trypan blue assay (see S6, ESI†). After the insertion by a long-taper nanopipette, the cells appear colorless within 60 min. In contrast, short-taper nanopipette inserted cells often show a distinctive blue color within 2 min. Thus, we are able to perform the live-cell experiments in an hour using the long-taper nanopipette. By using the nanopipette nanoprobe, pHi of HeLa and fibroblast cells in 1xPBS were measured based on the SERS spectra of 4-MBA molecule (results are shown in Fig. S7, ESI†). Based on the calibration curve of each nanopipette, the measured pHi values are always is in the range between 7.1 and 7.5, which are typical for the healthy cell in the culture environment.
Fig. 4.
(A) Bright-field images of HeLa cell before, during, and after the nanopipette-based nanoprobe insertion. (B) Normalized SERS spectra of the nanopipette-based nanoprobe in a fixed and live Fibroblast cell under pHe = 7.4 (blue color filled 1400 cm−1) and pHe = 6.0 (red color filled 1400 cm−1) stimulation for different time periods. (C) Variation of the pHi value in fixed Fibroblast cells, live Fibroblast and HeLa cells versus the time. The green shaded regions highlight the solution exchange time when the pHe is changed from 7.4 to 6.0. The error bars are obtained from the standard deviation of three separate measurements. The pH calibration curves for nanopipettes nanoprobes are shown in Fig. S9, ESI†.
Then we measured the dynamic pHi change induced by the change of pHe. All the experiments have been repeated at least three times with a good reproducibility. Fig. 4B shows a series of SERS spectra collected by the nanopipette tip inside the fixed (left) and live (right) Fibroblast cells at different times. The pHe of the medium was initially at 7.4 and replaced by pHe 6.0 medium during 4–6 min. Raman spectrum acquisition was interrupted for 2 min due to the solution exchange. Correspondingly, the magnitude of the peak at 1400 cm−1 in the SERS spectra is higher (in blue color) in the first 4 min and then drops at 6 min (in red color) for both fixed and live fibroblast cells. However, at a later time, the peak height at 1400 cm−1 increases gradually for live Fibroblast cells but not for the fixed Fibroblast.
Based on the calibration curve, we plotted the pHi change as a function of time for both cells in Fig. 4C. For the fixed Fibroblast cells, the pHi quickly drops and stays near 6.1. This is attributed to the increased permeability of the cell membrane of the fixed cell. The pHi of the live fibroblast cell also drops immediately with the change of pHe. However, the pHi of the live fibroblast cells gradually returns to ~7.3 after about 10 min while the cells are remained in the weak acidic medium with pHe 6. We did not observe visible changes in the morphology of the fibroblast cell. Therefore, the fibroblast cells likely adapt to environmental pH changes after 10 min. After that, the live fibroblast cells can maintain their cytoplasmic pH near neutral while they are still in a slightly acidic environment.
For comparison, we also studied the pHi response of HeLa cells when they were exposed to the same weak acidic environment. The result is shown at the right panel in Fig. 4C. Simiar to the live fibroblast cell results, the pHi of Hela cell decreases within 2 min with the exposure to the weak acidic environment. This change is much faster than the reported time of more than 10 min.19, 43 We speculate that the seal between cell membrane and nanopipette tip maybe affected by the sudden change of pHe and allows more protons to enter the cytoplasm, leading to the relatively fast drop of pHi.44 However, the leakage must be very small and can be resealed qucikly because the cells are still viable. Different from the live fibroblast cells, the pHi of HeLa cells begains to increase right after the drop and returns to the initial neutral value in about 6 min. The quick response and faster recovery of pHi of the HeLa cell indicates that the cancer cells may better regulate its pHi and adapt to the acidic environment. Although more experiments on different type of cancer cells are needed to further comfirm this point, the result is consistent with the fact that tumor is often in an acidic microenvironment.
Conclusions
In summary, we have successfully prepared SERS-active flexible nanopipettes for live-cell intracellular pH sensing. Because of the nanoscale size of the apex and the long taper geometry of the tip, the nanopipette is highly flexible and causes minimal cellular damage when it is inserted into the cell. The optimized AuNPs surface distribution and density on the outer surface of the glass nanopipette enable sensitive and fast responses in the SERS sgnal of pH-reporter molecule 4-MBA. The pH response of the SERS nanoprobe is linear in the dyanmic range between pH 6.0 and 8.0. The ratiometric SERS signal acquired from the nanopipette tip inside the cell is stable over time and the tip-to-tip variation is small. The non-specific adsorption of molecules and proteins on the AuNP surface only affects the overall Raman intensity but not the intensity ratio I1400/I1077 of 4-MBA molecule. We found that the cancerous HeLa cell could effectively regulate its pHi and better adapt to the weakly acidic extracellular environment than normal cells, such as fibroblast cells.
The intracellular pH sensing performance of the developed SERS-active nanopipette is comparable with the commercial available fluorescent probes such as pHrodo Red and pHrodo Green in terms of sensitivity (~0.2 pH units) but has the advantage of high spatial resolution, long-term stability and multiplex detection. There are still a lot of rooms to improve this sensor. The shape and size of the AuNP can be further optimized to improve the sensitivity. By introducing antifouling reagents, e.g., Polyethylene glycol (PEG), during chemical modification, the stability and selectivity of the sensor can also be improved. The pH reporter molecue 4-MBA can be repalced by or mixed with other molecules to expand the dyanmic range of the pH sensing. This sensor can also be easily tailored to detect other biomarkers by using different Raman probe molecules. The demonstrated on-demand local delivery is another advantage of this sensor. Therefore, we expect that the plasmonic nanopipette can be a promising sensor platform for various single-cell analysis applications, especially real-time intracellular sensing.
Supplementary Material
Acknowledgements
This work is supported by the Engineering Research Centers Program of the National Science Foundation under NSF Cooperative Agreement No. EEC-1647837 and NSF (CBET1454544). J. Guo thanks for the support provided by the Dissertation Year Fellowship award program. J. Moscoso acknowledges the REU support from the Science and Technology Center on Real-Time Functional Imaging, a National Science Foundation Science and Technology Center (grant DMR 1548924). Y. Lai and Y. Liu are supported by the National Institute of Health grant NIHR01ES023569. We also acknowledge FIU AMERI for the use of SEM.
Footnotes
Conflicts of interest
There are no conflicts to declare.
Electronic Supplementary Information (ESI) available: See DOI: 10.1039/x0xx00000x
Notes and references
- 1.Dittrich P and Jakubowski N, Analytical and Bioanalytical Chemistry, 2014, 406, 6957–6961. [DOI] [PubMed] [Google Scholar]
- 2.Yuan G-C, Cai L, Elowitz M, Enver T, Fan G, Guo G, Irizarry R, Kharchenko P, Kim J, Orkin S, Quackenbush J, Saadatpour A, Schroeder T, Shivdasani R and Tirosh I, Genome Biology, 2017, 18, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weiss S, Science, 1999, 283, 1676–1683. [DOI] [PubMed] [Google Scholar]
- 4.Okabe K, Inada N, Gota C, Harada Y, Funatsu T and Uchiyama S, Nature Communications, 2012, 3, 705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qian W, Huang X, Kang B and El-Sayed M, Journal of Biomedical Optics, 2010, 15, 046025. [DOI] [PubMed] [Google Scholar]
- 6.Willets KA, Analytical and Bioanalytical Chemistry, 2009, 394, 85–94. [DOI] [PubMed] [Google Scholar]
- 7.Ying Y-L, Hu Y-X, Gao R, Yu R-J, Gu Z, Lee LP and Long Y-T, Journal of the American Chemical Society, 2018, 140, 5385–5392. [DOI] [PubMed] [Google Scholar]
- 8.Teng KW, Ishitsuka Y, Ren P, Youn Y, Deng X, Ge P, Lee SH, Belmont AS and Selvin PR, eLife, 2016, 5, e20378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Llopis J, McCaffery JM, Miyawaki A, Farquhar MG and Tsien RY, Proceedings of the National Academy of Sciences, 1998, 95, 6803–6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.BERNAS T, ZARȨBSKI M, COOK RR and DOBRUCKI JW, Journal of Microscopy, 2004, 215, 281–296. [DOI] [PubMed] [Google Scholar]
- 11.Wright A, Bubb WA, Hawkins CL and Davies MJ, Photochemistry and Photobiology, 2002, 76, 35–46. [DOI] [PubMed] [Google Scholar]
- 12.Song L, van Gijlswijk RPM, Young IT and Tanke HJ, Cytometry, 1997, 27, 213–223. [PubMed] [Google Scholar]
- 13.Nolan JP, Duggan E, Liu E, Condello D, Dave I and Stoner SA, Methods, 2012, 57, 272–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kneipp K, Haka AS, Kneipp H, Badizadegan K, Yoshizawa N, Boone C, Shafer-Peltier KE, Motz JT, Dasari RR and Feld MS, Applied Spectroscopy, 2002, 56, 150–154. [Google Scholar]
- 15.Nabiev IR, Morjani H and Manfait M, European Biophysics Journal, 1991, 19, 311–316. [DOI] [PubMed] [Google Scholar]
- 16.Niu JJ, Schrlau MG, Friedman G and Gogotsi Y, Small, 2011, 7, 540–545. [DOI] [PubMed] [Google Scholar]
- 17.Singhal R, Orynbayeva Z, Kalyana Sundaram RV, Niu JJ, Bhattacharyya S, Vitol EA, Schrlau MG, Papazoglou ES, Friedman G and Gogotsi Y, Nature Nanotechnology, 2010, 6, 57. [DOI] [PubMed] [Google Scholar]
- 18.Scaffidi JP, Gregas MK, Seewaldt V and Vo-Dinh T, Analytical and Bioanalytical Chemistry, 2009, 393, 1135–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas RC, The Journal of physiology, 1974, 238, 159–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Özel RE, Lohith A, Mak WH and Pourmand N, RSC Advances, 2015, 5, 52436–52443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hailing L, Qiucen J, Jie P, Zeyu J, Jiao C, Lina J, Xinghua X and Kang W, Advanced Functional Materials, 2018, 28, 1703847. [Google Scholar]
- 22.Actis P, Maalouf MM, Kim HJ, Lohith A, Vilozny B, Seger RA and Pourmand N, ACS Nano, 2014, 8, 546–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jayant K, Wenzel M, Bando Y, Hamm JP, Mandriota N, Rabinowitz JH, Plante IJ-L, Owen JS, Sahin O, Shepard KL and Yuste R, Cell Reports, 2019, 26, 266–278.e265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang W, Zhao F, Li M, Zhang C, Shao Y and Tian Y, Angewandte Chemie International Edition, 2019, 58, 5256–5260. [DOI] [PubMed] [Google Scholar]
- 25.Vitol EA, Orynbayeva Z, Bouchard MJ, Azizkhan-Clifford J, Friedman G and Gogotsi Y, ACS Nano, 2009, 3, 3529–3536. [DOI] [PubMed] [Google Scholar]
- 26.Hanif S, Liu H-L, Ahmed SA, Yang J-M, Zhou Y, Pang J, Ji L-N, Xia X-H and Wang K, Analytical Chemistry, 2017, 89, 9911–9917. [DOI] [PubMed] [Google Scholar]
- 27.Freedman KJ, Crick CR, Albella P, Barik A, Ivanov AP, Maier SA, Oh S-H and Edel JB, ACS Photonics, 2016, 3, 1036–1044. [Google Scholar]
- 28.Hanif S, Liu H, Chen M, Muhammad P, Zhou Y, Cao J, Ahmed SA, Xu J, Xia X, Chen H and Wang K, Analytical Chemistry, 2017, 89, 2522–2530. [DOI] [PubMed] [Google Scholar]
- 29.Masson J-F, Breault-Turcot J, Faid R, Poirier-Richard H-P, Yockell-Lelièvre H, Lussier F and Spatz JP, Analytical Chemistry, 2014, 86, 8998–9005. [DOI] [PubMed] [Google Scholar]
- 30.Liu H-L, Cao J, Hanif S, Yuan C, Pang J, Levicky R, Xia X-H and Wang K, Analytical Chemistry, 2017, 89, 10407–10413. [DOI] [PubMed] [Google Scholar]
- 31.Casey JR, Grinstein S and Orlowski J, Nature Reviews Molecular Cell Biology, 2010, 11, 50–61. [DOI] [PubMed] [Google Scholar]
- 32.Han J and Burgess K, Chemical Reviews, 2010, 110, 2709–2728. [DOI] [PubMed] [Google Scholar]
- 33.Guo J, Pan J, Chang S, Wang X, Kong N, Yang W and He J, Small, 2018, 14, 1704164. [DOI] [PubMed] [Google Scholar]
- 34.Laffafian I and Hallett MB, Biophysical Journal, 1998, 75, 2558–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schrlau MG and Bau HH, 2008. [Google Scholar]
- 36.Solís DM, Taboada JM, Obelleiro F, Liz-Marzán LM and García de Abajo FJ, ACS Photonics, 2017, 4, 329–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma T, Guo J, Chang S, Wang X, Zhou J, Liang F and He J, Physical Chemistry Chemical Physics, 2019, 21, 15940–15948. [DOI] [PubMed] [Google Scholar]
- 38.Zong Y, Guo Q, Xu M, Yuan Y, Gu R and Yao J, RSC Advances, 2014, 4, 31810–31816. [Google Scholar]
- 39.Handa S, Yu Y and Futamata M, Vibrational Spectroscopy, 2014, 72, 128–133. [Google Scholar]
- 40.Phan HT and Haes AJ, The Journal of Physical Chemistry C, 2018, 122, 14846–14856. [Google Scholar]
- 41.Lu G, Shrestha B and Haes AJ, The Journal of Physical Chemistry C, 2016, 120, 20759–20767. [Google Scholar]
- 42.Lee SB, Kim K and Kim MS, Journal of Raman Spectroscopy, 1991, 22, 811–817. [Google Scholar]
- 43.Fellenz MP and Gerweck LE, Radiation Research, 1988, 116, 305–312. [PubMed] [Google Scholar]
- 44.Angelova MI, Bitbol A-F, Seigneuret M, Staneva G, Kodama A, Sakuma Y, Kawakatsu T, Imai M and Puff N, Biochimica et Biophysica Acta (BBA) - Biomembranes, 2018, 1860, 2042–2063. [DOI] [PubMed] [Google Scholar]
- 45.Kato Y, Ozawa S, Miyamoto C, Maehata Y, Suzuki A, Maeda T and Baba Y, Cancer Cell Int, 2013, 13, 89–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Webb BA, Chimenti M, Jacobson MP and Barber DL, Nature Reviews Cancer, 2011, 11, 671–677. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.