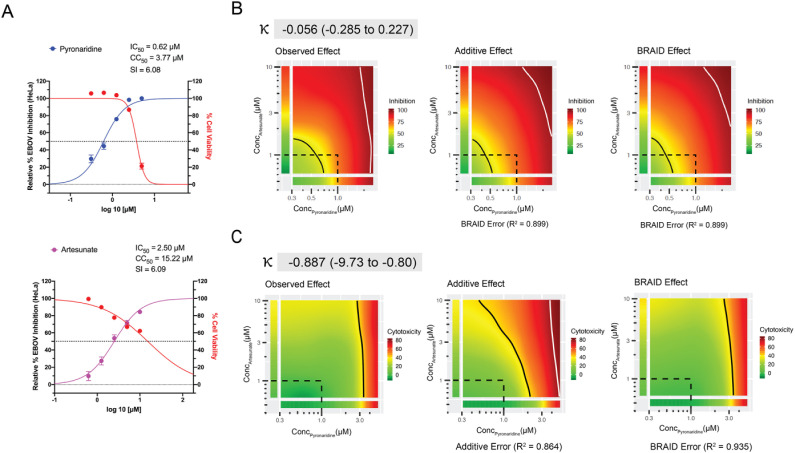
Fig. 3.
Combination data for the pyronaridine and artesunate checkerboard assay in HeLa cells. A) Inhibition/cytotoxicity plots for the pyronaridine and artesunate controls (compound tested in the absence of the other compound). Controls were run in triplicate at 5 concentrations per plate, so the total number from replicates for each compound varied (Pyronaridine, n = 27; Artesunate, n = 18). Error bars represent the SEM at each concentration tested. B) Graphical representations (from left to right) of the inhibition plots of the smoothed raw data, predicted additive inhibition and predicted inhibition using the 7-parameter BRAID analysis. It is noted that inhibition data under toxic concentrations (>50% cell death) were removed from the analysis. The “Additive” or “BRAID” error represents the corresponding accuracy of fit with the “Observed Effect”. κ represents the combinatory effect where κ = 0 implies additivity, and κ > 0 implies synergy, “strong synergy” corresponds to κ = 2.5, “mild synergy” corresponds to κ = 1, “mild antagonism” corresponds to κ = −0.66, and “strong antagonism” corresponds to κ = −1. C) Representation of the cytotoxicity (toxicity is representative of % cell death from control) arranged in the same manner as inhibition.