Figure 1.
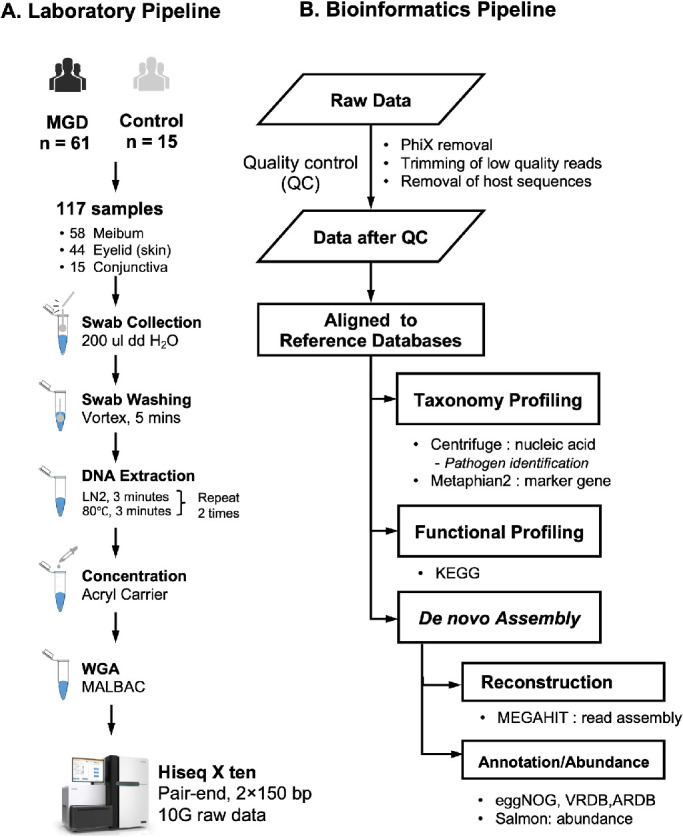
Study design. (A) Laboratory Pipeline. Swabs were collected from 76 subjects, including 61 MGD patients and 15 HC volunteers. Totally, 117 samples had qualified sequencing results (Method), including 58 were from meibum, 44 from skin of eyelid, and 15 from the conjunctiva. Liquid nitrogen (LN) was used for DNA extraction, and Acryl carrier was used to enrich the DNA fragment and decrease the volumes of DNA solution. Whole-genome amplification (WGA) was applied to obtain an adequate amount of DNA for sequencing library construction. (B) Bioinformatic Pipeline. Quality control process low quality reads, PhiX, and host sequences were removed in the quality control process. Multiple tools and public databases were applied to annotate sequencing fragments and de novo assembly.
