Extended Data Fig. 4. AKAP95 phase separation in vitro.
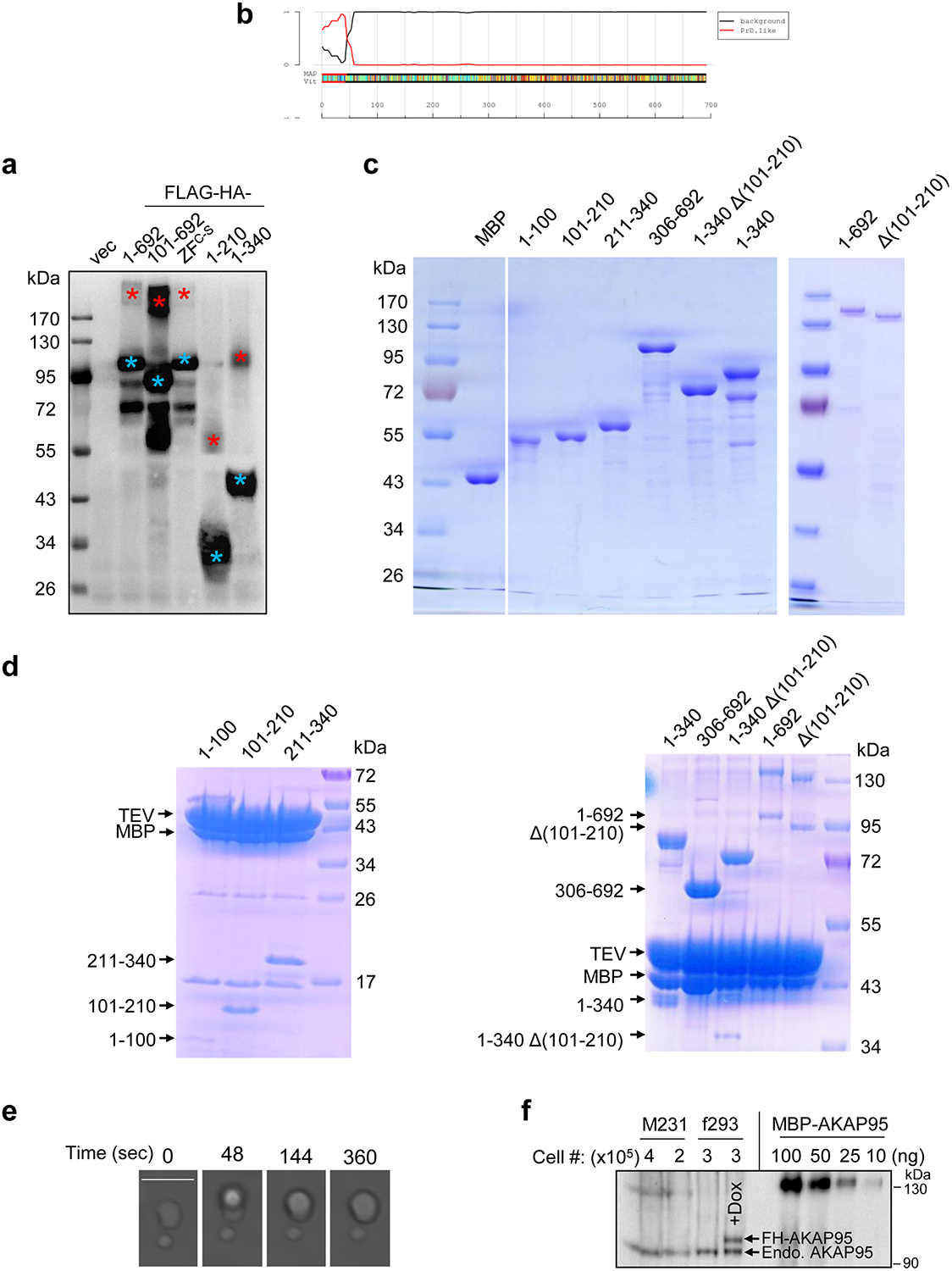
a, 293T cells were transfected with either empty vector (vec), or indicated AKAP95 construct with FLAG-HA-tag. Following α-Flag IP, the pulldown proteins were boiled and resolved by SDS-PAGE and detected by immunoblotting with α-HA. Blue and red asterisks indicate monomer and dimer, respectively. b, Identification of 1–100 as a probable prion subsequence on AKAP95. By the PLAAC program, using homo sapiens as background and core length of 30. c, Purified MBP and MBP fused to AKAP95 truncations as indicated or full-length AKAP95 (1–692) were resolved on SDS-PAGE and stained with Coomassie blue. d, MBP fused to AKAP95 truncations as indicated or full-length AKAP95 were resolved on SDS-PAGE and stained with Coomassie blue following treatment with TEV protease. Note that the cleaved MBP serves as a better indicator for cleavage efficiency as staining signal various for protein fragments of different sequences and sizes. e, Another event of fusion of two droplets formed by 50 μM MBP-AKAP95 (101–210) in 30 mM NaCl and 10% of PEG6000 after treatment with TEV protease for 30 min. Scale bar, 5 μm. Also see Supplementary Video 1. f, Quantification of nuclear AKAP95 concentration by anti-AKAP95 Western blot. Total lysates from indicated number of MDA-MB-231 (M231) and flp-TREx 293 cells (f293, un-induced and dox-induced for FH-AKAP95 expression) were loaded, along with indicated ng of purified MBP-AKAP95. AKAP95 signal of un-induced f293 is similar to that of 25 ng of MBP-AKAP95. All experiments were repeated 2 times. Uncropped blots are provided as in source data extended data fig 4.
