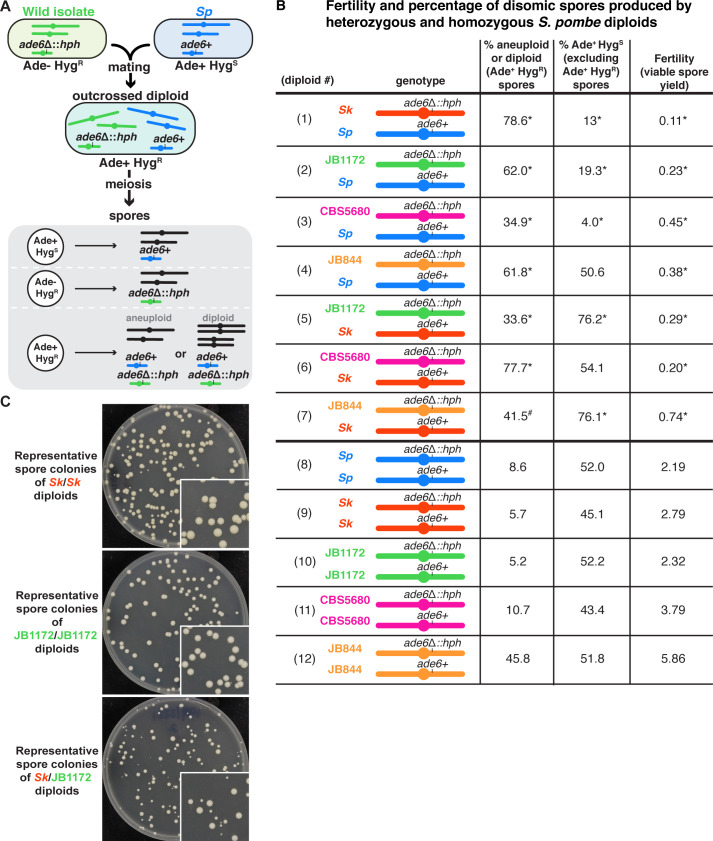
Figure 1. Outcrossed S. pombe diploids frequently produce disomic spores.
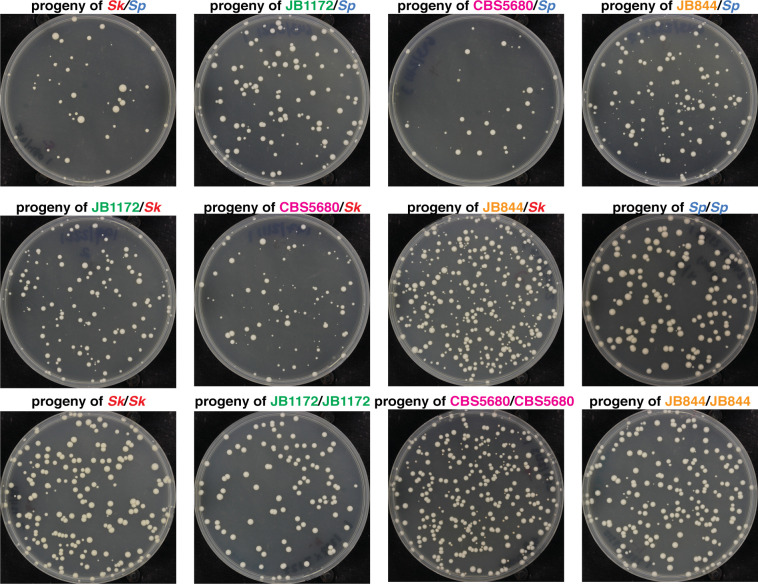
(A) Schematic of the experimental approach. The ade6 gene is linked to centromere 3. The karyotypes of JB1172, CBS5680, and JB844 are unknown, but outside of an inversion on Sp chromosome 1, Sp likely represents the ancestral karyotype (Avelar et al., 2013; Brown et al., 2011). (B) Phenotypes of heterozygous or homozygous S. pombe diploids. Allele transmission of chromosome 3 was assayed using co-dominant markers at ade6 (ade6+ and ade6Δ::hphMX6). The ade6+ allele confers an Ade+ phenotype, while the ade6Δ::hphMX6 provides resistance to Hygromycin B (HygR). Heterozygous aneuploid or diploid spores are Ade+ HygR. The fertility was measured using the viable spore yield assay to determine the number of viable spores per viable diploid. In the absence of drive, we expect 50% of the spores to be Ade+ HygS. A significant departure from 50% indicates drive favoring the overrepresented allele. The phenotypes of each heterozygote were compared to those of homozygous diploids from both parental strain backgrounds. * indicates p-value<0.025 (G-test [Ade+ HygR spores] and Wilcoxon test [fertility]) for the heterozygotes relative to the homozygous diploids from both parental backgrounds. Diploid 7 was only significantly different (p-value<0.025) in the frequency of Ade+ HygR spores when compared to diploid 9, but not when compared to diploid 12. This is indicated with #. To detect biased allele transmission (Ade+ HygS), diploids 2–4 were compared to diploid 8 and diploids 5–7 were compared to diploid 9. * indicates p-value<0.05 (G-test [allele transmission]). More than 200 viable spores were scored for each diploid. Raw data can be found in Figure 1—source data 1 and Figure 1—source data 2. (C) Representative images of the viable spore colonies generated by homozygous Sk and JB1172 diploids and heterozygous Sk/JB1172 diploids. Images of colonies generated by other diploids are shown in Figure 1—figure supplement 2.