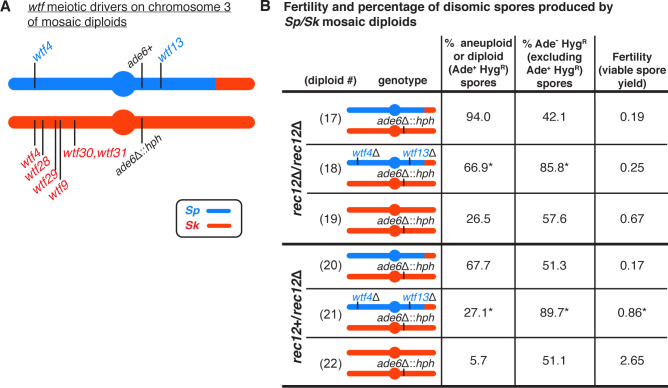
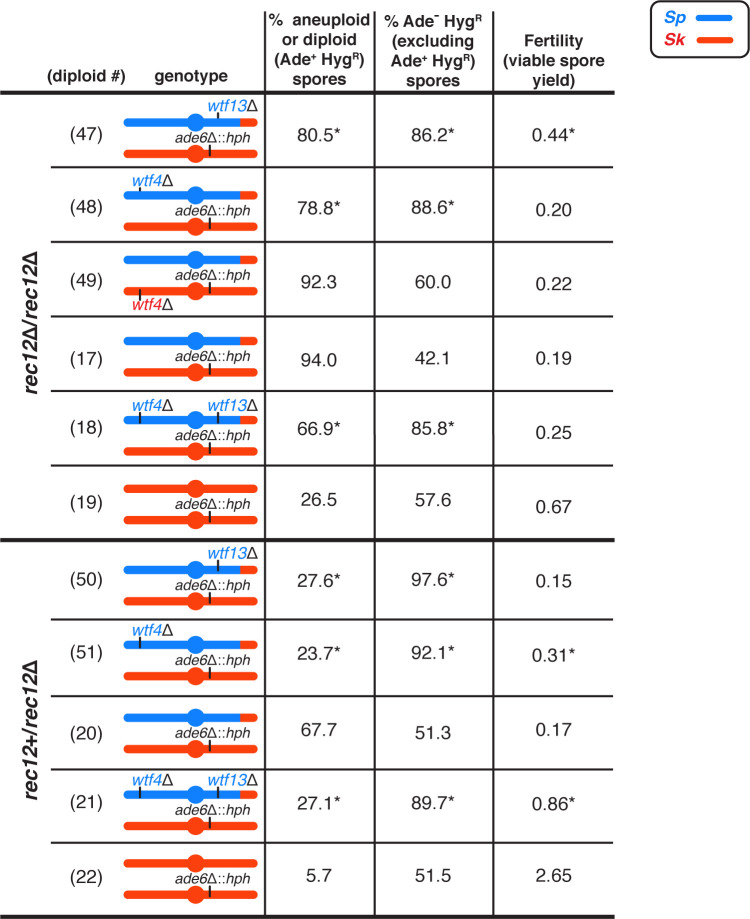
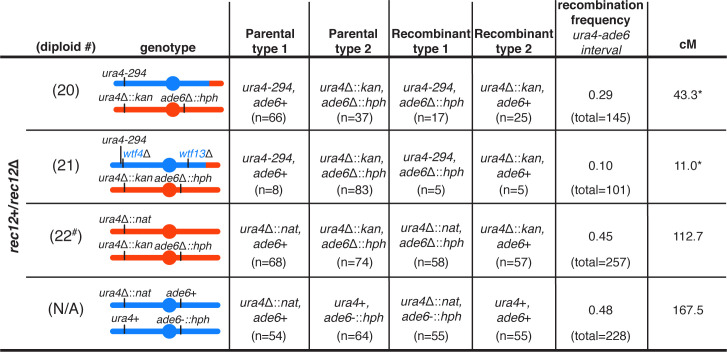
Figure 3. wtf meiotic driver competition contributes to the high disomy in spores produced by Sp/Sk mosaic diploids.
(A) Schematic of the predicted wtf meiotic drivers found on chromosome 3 of the Sp/Sk mosaic diploid. Sp-derived DNA is depicted in blue and Sk-derived DNA in red. (B) Phenotypes of mosaic and control diploids in rec12∆/rec12∆ and rec12+/rec12∆ backgrounds. Allele transmission of chromosome 3 was assayed using markers at ade6 (linked to centromere 3). In the absence of drive, we expect 50% of the spores to be Ade- HygR. Any significant deviation from the expected 50% indicates drive favoring the overrepresented allele. To determine the contribution of wtf meiotic drivers to the frequency of disomic spores and fertility, diploid 18 was compared to diploid 17, and diploid 21 was compared to diploid 20. To determine if there was biased allele transmission, diploids 17 and 18 were compared to control diploid 19, and diploids 20 and 21 were compared to control diploid 22. More than 300 viable spores were scored for each diploid. * indicates p-value<0.05 (G-test [allele transmission and Ade+ HygR spores] and Wilcoxon test [fertility]). Raw data can be found in Figure 3—source data 1 and Figure 3—source data 2.