Figure 2.
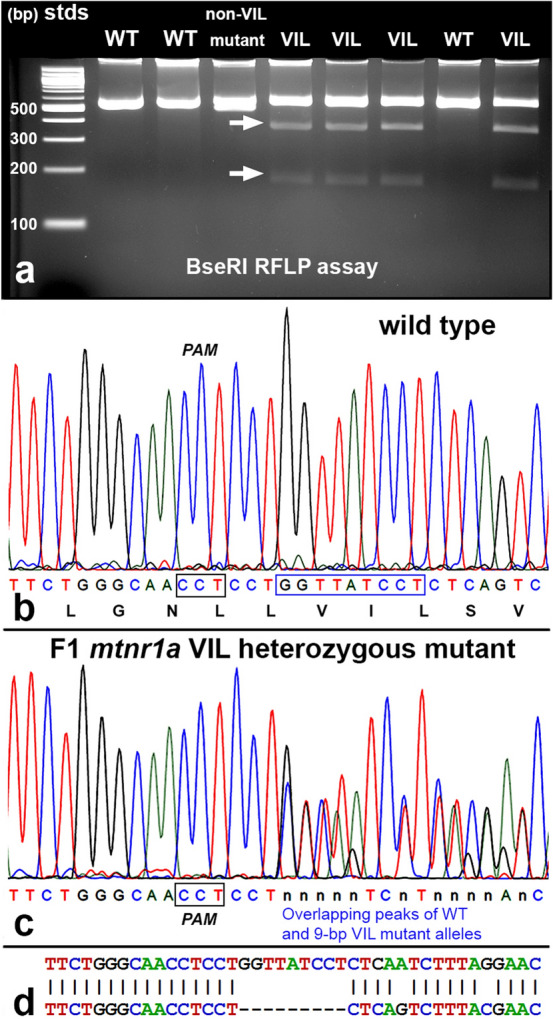
Identification of heterozygous F1 animals with CRISPR/Cas9 indel mutations of the mtnr1a gene. (a) Representative RFLP analysis shows that PCR products of mtnr1a VIL heterozygotes display two predicted cleavage bands (arrows), whereas WT and non-VIL mutant DNA is not cleaved. The creation of a BseRI site by the VIL deletion is illustrated in Supplementary Fig. S3. (b) Direct PCR Sanger sequencing chromatogram of F1 WT progeny display prominent peaks for all bases in the T2 sgRNA target area. The T2 PAM sequence (CCT) is boxed in black, and the downstream 9-bp sequence of the mtnr1a VIL deletion mutation is boxed in blue. The corresponding amino acid sequence is shown below the nucleotide codon sequence. (c) In an F1 tadpole heterozygous for the mtnr1a 9-bp (VIL) deletion, the trace decomposes (i.e., mixed base calls) at the predicted target site 4-bp downstream of the T2 PAM site, and continues to the end of the trace, as expected for a heterozygous indel mutation. The decomposed sequence represents the overlapping peaks of the mtnr1a WT and VIL alleles. (d) Partial view of alignments from Poly Peak Parser (https://yosttools.genetics.utah.edu/PolyPeakParser/) output revealing the heterozygous 9-bp mtnr1a VIL deletion mutation. The WT allele is the top line and the mutant allele is the bottom line.
