Abstract
The novel coronavirus SARS‐CoV‐2 is the causative agent of the global coronavirus disease 2019 (COVID‐19) outbreak. In addition to pneumonia, other COVID‐19‐associated symptoms have been reported, including loss of smell (anosmia). However, the connection between infection with coronavirus and anosmia remains enigmatic. It has been reported that defects in olfactory cilia lead to anosmia. In this Viewpoint, we summarize transmission electron microscopic studies of cilia in virus‐infected cells. In the human nasal epithelium, coronavirus infects the ciliated cells and causes deciliation. Research has shown that viruses such as influenza and Sendai attach to the ciliary membrane. The Sendai virus enters cilia by fusing its viral membrane with the ciliary membrane. A recent study on SARS‐CoV‐2–human protein–protein interactions revealed that the viral nonstructural protein Nsp13 interacts with the centrosome components, providing a potential molecular link. The mucociliary escalator removes inhaled pathogenic particles and functions as the first line of protection mechanism against viral infection in the human airway. Thus, future investigation into the virus–cilium interface will help further the battle against COVID‐19.
Keywords: COVID‐19, SARS‐CoV‐2, smell loss, cilia
The connection between coronavirus infection and anosmia remains enigmatic. Defects in olfactory cilia lead to anosmia. This viewpoint summarizes transmission electron microscopic studies of cilia in virus‐infected cells: Coronavirus infects the ciliated cells in human nasal epithelium and causes deciliation. Influenza viruses attach to the ciliary membrane, and Sendai virus enters cilia by fusing its viral membrane with the ciliary membrane.
Abbreviations
- ACE2
angiotensin‐converting enzyme 2
- COVID‐19
coronavirus disease 2019
- MERS‐CoV
Middle East respiratory syndrome coronavirus
- SARS‐CoV
severe acute respiratory syndrome coronavirus
- SARS‐CoV‐2
severe acute respiratory syndrome coronavirus 2
- scRNA‐seq
single‐cell RNA sequencing
- TEM
transmission electron microscopy
COVID‐19 impairs smell
A novel coronavirus designated as severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is an enveloped virus of the family Coronaviridae with a positive‐sense, single‐stranded RNA genome [1, 2]. SARS‐CoV‐2 was identified as the etiological agent that causes the global outbreak of coronavirus disease 2019 (COVID‐19) [1, 2]. Historically, several mild common cold coronaviruses such as hCoV‐OC43 were reported to infect humans [3]. However, highly pathogenic human coronaviruses have emerged during the past two decades, including SARS‐CoV in 2002 and MERS‐CoV in 2012, which can result in acute respiratory distress syndrome, potentially leading to a reduction in lung function or death [2, 4, 5]. Despite the low fatality rate, SARS‐CoV‐2 spreads more efficiently than SARS‐CoV or MERS‐CoV, which makes it challenging to contain [5, 6]. By late June 2020, more than 9.4 million laboratory‐confirmed infections were reported worldwide, including 0.48 million deaths [7]. The active therapeutic measures available to counteract the SARS‐CoV‐2 virus are limited, and the associated COVID‐19 pathology is not well documented. Thus, it is urgent to understand how SARS‐CoV‐2 hijacks the host during infection and how the disease disrupts the cellular structure and function.
Coronavirus disease 2019 is predominantly a lung infection, causing cough, fever, and fatigue, but other symptoms have been reported throughout the body [1, 8]. A previously unrecognized symptom of SARS‐CoV‐2 disease, loss of smell, has been acknowledged by clinicians and the public globally. A recent clinical study described that smell and taste impairments were strongly associated with COVID‐19 patients in the United States [9]. A similar correlation between the new‐onset chemosensory dysfunction (i.e., smell and taste loss) and COVID‐19 positivity was reported from Iran and Italy since the COVID‐19 outbreak, suggesting that loss of smell or taste may be considered as subclinical markers or a potential early symptom [10, 11]. Patients with the new‐onset anosmia may be asymptomatic carriers of SARS‐CoV‐2 infection who require self‐isolation; otherwise, they risk facilitating the spread of disease [9, 12]. Indeed, the Centers for Disease Control and Prevention added ‘loss of taste or smell’ as one of six new symptoms of COVID‐19 on their advisory page on April 27, 2020 [13]. However, the pathological evidence for connecting COVID‐19 infection with anosmia is lacking.
The current theory of smell mechanism posits that numerous odorant receptors, which accumulate on sensory cilia in the olfactory epithelium, perceive the corresponding odorants and transduce the signal to the olfactory cortex in the brain [14]. Primary cilia are microtubule‐based organelles that are projected from the surface of the cell, functioning as the cell's antenna to perceive various environmental stimuli [15]. Ciliary dysfunction leads to more than 35 types of diseases, which are collectively termed ciliopathies [15]. The olfaction of humans and many other species requires the standard structure and function of olfactory cilia, and anosmia turns out to be a common feature of some types of ciliopathies such as Bardet–Biedl syndrome [15]. While the viral‐induced inflammation can perturb the nervous system and cause smell loss, viral infection may disrupt the ciliary structure, which abolishes the ciliary localization of olfactory receptors, thereby preventing the perception of odorant molecules. Here, we review the early transmission electron microscopic (TEM) results of the ciliary ultrastructure during viral infection, in the light of the latest molecular advancements.
Cilia in virus‐infected cells
Firstly, coronavirus infects the ciliated cells in the human nasal epithelium, resulting in the loss of cilia. In 1994, Bjorn Afzelius, who first described primary ciliary dyskinesia, reported the ultrastructural study of the nasal mucosa from a patient diagnosed with chronic rhinitis and bronchitis [16]. Afzelius detected coronavirus in the ciliated cells (Fig. 1C). From the SARS‐CoV‐2‐infected macaques, the latest pathological studies found the expression of SARS‐CoV‐2 antigens in the ciliated nasal epithelial cells [17]. Coronaviruses enter the host cells using the spike (S) protein, which binds to a specific cellular receptor, followed by protease‐dependent S protein priming. Both SARS‐CoV and SARS‐CoV‐2 employ the angiotensin‐converting enzyme 2 (ACE2) as the host receptor and TMPRSS2 as the cellular protease. Single‐cell RNA sequencing (scRNA‐seq) datasets from the Human Cell Atlas consortium described the expression of the receptor and protease in nasal epithelial cells [18, 19], providing the molecular basis for coronavirus entry into the human nasal epithelia.
Fig. 1.
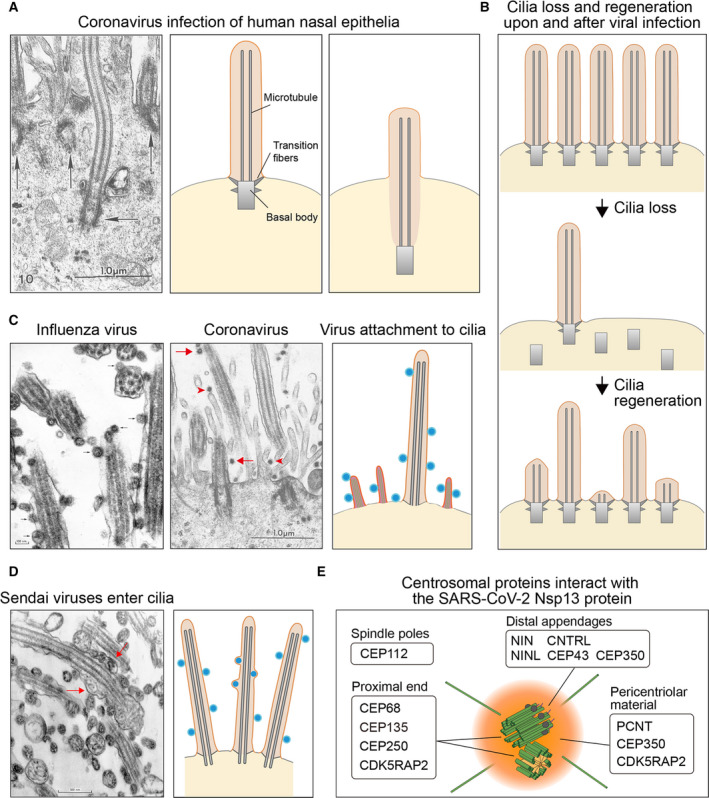
(A) Left: The electron microscopy image from Ref. [16] shows the sunk cilium and basal body (horizontal arrow) in a ciliated human nasal epithelial cell. The neighbor basal bodies remain at original places (vertical arrows). Scale bar: 1 μm. Middle: schematic diagram of the primary cilium. Right: schematic diagram of the sunk cilium and basal body. (B) Loss and regeneration of cilia during virus infection. (C) Left: Influenza viruses attach to the cilia of trachea epithelial cells (arrows) [23]. Scale bar: 100 nm. Middle: Coronaviruses attach to the cilia (arrows) and microvilli (arrowheads) of nasal epithelial cells [16]. Scale bar: 1 μm. Right: diagram of cilia and microvilli with virus particles attached. (D) Left: Sendai viruses adhere to the cilia of trachea epithelial cells [23]. Some viruses fuse the viral membrane with the cilia membrane, and the nucleocapsids enter the cilia (arrows). Scale bar: 500 nm. Right: A diagram shows that viruses attach to and fuse with the cilia membrane. (E) The Nsp13 protein in SARS‐CoV‐2 interacts with the centrosome proteins [24]. Figure reproduced from Ref. [16, 23].
Afzelius showed that, although the virions did not unduly damage the ciliated epithelium, the loss of cilia represented one of the most striking ultrastructural abnormalities in coronavirus‐infected cells (Fig. 1A). Two alternative models explain how a cell removes its primary cilium [20]. In the amputation model, the entire cilium is acutely eliminated at the ciliary base, such as the pH shock treatment removing Chlamydomonas flagella. In the absorption model, the cilium can be gradually absorbed by the cytoplasm. Afzelius reported that cilia retracted into the cell body and found the 9 + 2 axonemal doublet microtubules inside the cytoplasm, which suggests that viral infection causes deciliation through cilium absorption (Fig. 1A). In line with this model, the basal bodies from the deciliated cells sunk into the cytoplasm, which was 7–8 microns below the plasma membrane (Fig. 1A). The formation of the apical vesicles that contain virion particles allowed Afzelius to speculate further that these ectopic vesicles may cause the basal bodies to be displaced from the cell surface, leading to cilia retraction [16]. Importantly, loss of cilia is evident in SARS and other viral infections [21]. Correlatively, TEM studies detected the intermediate and short cilia that undergo regeneration in the course of patient recovery, including smell restoration (Fig. 1B) [22]. However, no direct research determined whether coronavirus infects olfactory neurons, olfactory epithelia, or nasal epithelia localized in the same nasal cavity, making the whole area equally at risk of infection.
Various viruses enter the host cells by directly fusing with the plasma membrane or through endocytosis, but relatively little is known about whether the viruses can enter the host cells from cilia by directly attaching to the ciliary membrane. TEM studies provide evidence that cilia can be the absorption site or entry site for viral infection. In 1970, Dourmashkin and Tyrrell showed that the influenza virus lined up along the ciliary membrane (Fig. 1C) [23]. Their TEM images convincingly showed that the Sendai virus not only attached to cilia but also fused to the viral membrane with the ciliary membrane (Fig. 1D). After entering the cilia, the Sendai virus nucleocapsid became partially uncoiled and spread between the ciliary membrane and axoneme [23]. Afzelius's TEM image also revealed the attachment of coronavirus to cilia in nasal epithelia (Fig. 1B) [16]. Thus, cilia serve as the absorption sites for different families of viruses, and, at least for the Sendai virus, cilia appear to be one of the entry sites for viral infection. These studies revealed that the viruses enter the host cell at multiple locations; for example, the viruses adhere to the plasma membrane of the microvilli [16, 23], another type of cell surface protrusion that is supported by actin bundles.
The recent research on the SARS‐CoV‐2–human protein–protein interactions revealed a potential molecular link between virus and cilium biology [24]. The SARS‐CoV‐2 30 kb genome encodes 14 Orfs. Among them, polyprotein Orf1a/Orf1ab is autoproteolytically processed into 16 nonstructural proteins. Except for Nsp3 and Nsp16, all the predicted 27 proteins were individually expressed in human HEK293T cells with an affinity tag. Affinity purification and mass spectrometry‐based proteomics systematically identified the binding partners of SARS‐CoV‐2 proteins in the host cells. Among the high‐confidence interactors, Nsp13 binds to as many as 12 components in the centrosome, including three in the pericentrosomal region, five in the distal appendage of the centriole, and four in the proximal end of the centriole (Fig. 1E) [25, 26, 27]. These physical interactions have not been experimentally validated, and the underlying biological relevance is unclear. We cannot exclude the possibility that the host cells treat the viral protein Nsp13 as cellular garbage by disposing it onto the centrosome, where it glues to the centrosome. Alternatively, Nsp13 may actively localize to the centriole and compete with the endogenous binding partners of the centrosome proteins, abolishing the physiological interactions within the structure. Therefore, the ectopic interactions of the SARS‐CoV‐2 protein with the host proteins may disrupt the centriolar structure or the interaction between centriole and cilia, which eventually leads to deciliation. While the RNA sequence of the coronaviruses from which Afzelius showed their ability to affect cilia structure is not available, Nsp13 is a highly conserved protein in coronaviruses, as evidenced by 100% amino acid sequence identity of Nsp13 in SARS‐CoV‐2 and SARS‐CoV. Because HEK293T cells do not develop cilia, a similar protein interaction network analysis will need to be performed in human ciliated cells, which may yield additional insights into the interactions between SARS‐CoV‐2 viral proteins and the centriolar or ciliary proteins. To directly examine the impact of Nsp13 in ciliogenesis, a future study will need to express Nsp13 in ciliated cells and monitor cilia integrity over time.
Outstanding questions
Despite the profound importance of the mucociliary clearance in removing inhaled viral pathogens, cilium biology and viral infection appear to have been two parallel fields for decades. The outstanding questions in regard to the virus–cilium interface include: How does viral infection cause deciliation? Can we block deciliation to preserve a functional mucociliary escalator for viral particle clearance? How does the virus enter cilia? ACE2 and TMPRSS2 are expressed in both nasal and bronchial epithelia [18, 19]. How does a virus move from cilia to cell body? Does the nucleocapsid hijack the retrograde intraflagellar transport dynein‐2 motor protein? How could we regenerate cilia during patient recovery? While the ancient TEM images, the latest scRNA‐seq, and proteomic studies provide clues, the molecular and cellular insights into these problems still await future investigations.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
WL and GO outlined the content and wrote the manuscript. ML prepared the figures.
Acknowledgements
This work was supported by the National Key R&D Program of China (2017YFA0102900) and the National Natural Science Foundation of China (31671444 and 31871352 to WL and 31991190 to GO).
References
- 1. Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R et al (2020) A novel coronavirus from patients with pneumonia in China, 2019. New Engl J Med 382, 727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N et al (2020) Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395, 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zumla A, Chan JF, Azhar EI, Hui DS & Yuen KY (2016) Coronaviruses ‐ drug discovery and therapeutic options. Nat Rev Drug Discov 15, 327–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guarner J (2020) Three emerging coronaviruses in two decades. Am J Clin Pathol 153, 420–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rabaan AA, Al‐Ahmed SH, Haque S, Sah R, Tiwari R, Malik YS, Dhama K, Yatoo MI, Bonilla‐Aldana DK & Rodriguez‐Morales AJ (2020) SARS‐CoV‐2, SARS‐CoV, and MERS‐COV: a comparative overview, Le infezioni in medicina. Infez Med 28, 174–184. [PubMed] [Google Scholar]
- 6. Rabi FA, Al Zoubi MS, Kasasbeh GA, Salameh DM & Al‐Nasser AD (2020) SARS‐CoV‐2 and coronavirus disease 2019: what we know so far. Pathogens 9, 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. World Health Organization (2020) Coronavirus Disease 2019 (COVID‐19) Pandemic, 2020. Available from: https://www.who.int/emergencies/diseases/novel‐coronavirus‐2019. [cited 2020 Jun, 24] [Google Scholar]
- 8. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X et al (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yan CH, Faraji F, Prajapati DP, Boone CE & DeConde AS (2020) Association of chemosensory dysfunction and Covid‐19 in patients presenting with influenza‐like symptoms. Int Forum Allergy Rhinol 10, 806–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bagheri SHR, Asghari AM, Farhadi M, Shamshiri AR, Kabir A, Kamrava SK, Jalessi M, Mohebbi A, Alizadeh R, Honarmand AA et al (2020) Coincidence of COVID‐19 epidemic and olfactory dysfunction outbreak. medRxiv. 2020.03.23.20041889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Giacomelli A, Pezzati L, Conti F, Bernacchia D, Siano M, Oreni L, Rusconi S, Gervasoni C, Ridolfo AL, Rizzardini G, et al (2020) Self‐reported olfactory and taste disorders in SARS‐CoV‐2 patients: a cross‐sectional study. Clin Infect Dis 71, 889–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pallanti S (2020) Importance of SARs‐Cov‐2 anosmia: from phenomenology to neurobiology. Compr Psychiatry 100, 152184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Centers for Disease Control and Prevention (2020) Symptoms of Coronavirus. Available from: https://www.cdc.gov/coronavirus/2019‐ncov/symptoms‐testing/symptoms.html [Google Scholar]
- 14. Glezer I & Malnic B (2019) Olfactory receptor function. Handb Clin Neurol 164, 67–78. [DOI] [PubMed] [Google Scholar]
- 15. Reiter JF & Leroux MR (2017) Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol 18, 533–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Afzelius BA (1994) Ultrastructure of human nasal epithelium during an episode of coronavirus infection. Virchows Arch 424, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rockx B, Kuiken T, Herfst S, Bestebroer T, Lamers MM, Oude Munnink BB, de Meulder D, van Amerongen G, van den Brand J, Okba NMA et al (2020) Comparative pathogenesis of COVID‐19, MERS, and SARS in a nonhuman primate model. Science 368, 1012–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lukassen S, Chua RL, Trefzer T, Kahn NC, Schneider MA, Muley T, Winter H, Meister M, Veith C, Boots AW et al (2020) SARS‐CoV‐2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J 39, e105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sungnak W, Huang N, Becavin C, Berg M, Queen R, Litvinukova M, Talavera‐Lopez C, Maatz H, Reichart D, Sampaziotis F et al (2020) SARS‐CoV‐2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med 26, 681–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mirvis M, Siemers KA, Nelson WJ & Stearns TP (2019) Primary cilium loss in mammalian cells occurs predominantly by whole‐cilium shedding. PLoS Biol 17, e3000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nicholls JM, Poon LL, Lee KC, Ng WF, Lai ST, Leung CY, Chu CM, Hui PK, Mak KL, Lim W et al (2003) Lung pathology of fatal severe acute respiratory syndrome. Lancet 361, 1773–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rautiainen M, Nuutinen J, Kiukaanniemi H & Collan Y (1992) Ultrastructural changes in human nasal cilia caused by the common cold and recovery of ciliated epithelium. Ann Otol Rhinol Laryngol 101, 982–987. [DOI] [PubMed] [Google Scholar]
- 23. Dourmashkin RR & Tyrrell DA (1970) Attachment of two myxoviruses to ciliated epithelial cells. J Gen Virol 9, 77–88. [DOI] [PubMed] [Google Scholar]
- 24. Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, O'Meara MJ, Rezelj VV, Guo JZ, Swaney DL et al (2020) A SARS‐CoV‐2 protein interaction map reveals targets for drug repurposing. Nature 583, 459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jakobsen L, Vanselow K, Skogs M, Toyoda Y, Lundberg E, Poser I, Falkenby LG, Bennetzen M, Westendorf J, Nigg EA et al (2011) Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods. EMBO J 30, 1520–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kumar A, Rajendran V, Sethumadhavan R & Purohit R (2013) CEP proteins: the knights of centrosome dynasty. Protoplasma 250, 965–983. [DOI] [PubMed] [Google Scholar]
- 27. Casenghi M, Meraldi P, Weinhart U, Duncan PI, Korner R & Nigg EA (2003) Polo‐like kinase 1 regulates Nlp, a centrosome protein involved in microtubule nucleation. Dev Cell 5, 113–125. [DOI] [PubMed] [Google Scholar]


