Fig. 4. Identification of a long terminal repeat (LTR) retrotransposon in the promoter region of MELO3C001864, a potential core regulator of fruit ripening.
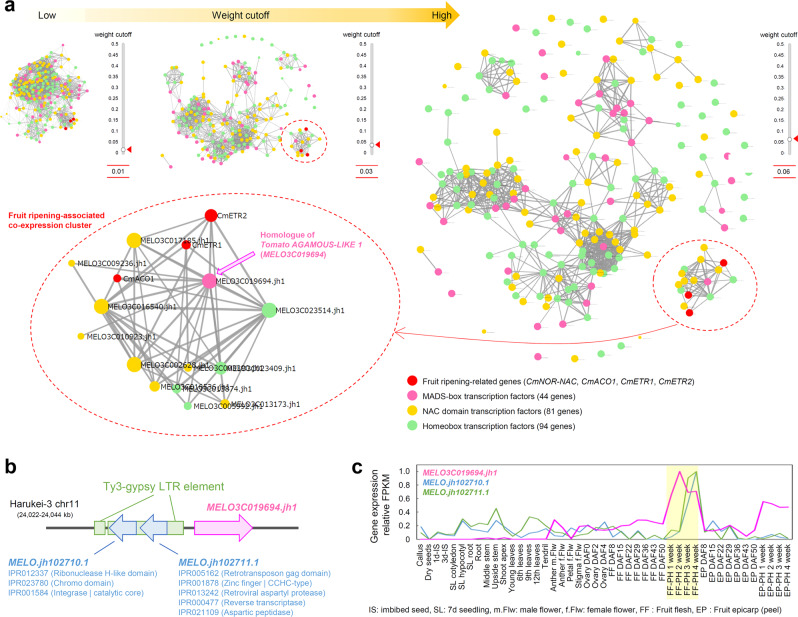
a Co-expression analysis of four known fruit-ripening-related genes with 81 NAC domain, 90 homeobox, and 42 MADS-box transcription factors based on the updated tissue-wide transcriptome dataset. In the updated version of “Co-expression viewer” in the Melonet-DB (https://melonet-db.dna.affrc.go.jp/ap/mds), the weight cutoff value is changeable in a real-time manner with the slider bar function present in the interface window. At weight cutoff = 0.06, one co-expression cluster, including known fruit-ripening-related genes, is detached from other clusters (shown by dashed red circles). A melon homologue of Tomato AGAMOUS-LIKE 1 (TAGL1), MELO3C019694, is positioned at the center of this fruit-ripening-associating co-expression cluster (indicated by pink-filled arrow). b Schematic illustration of the genomic region around MELO3C019694. Two long terminal repeat (LTR)/gypsy elements (1956 bp and 6941 bp) are identified by RepeatMasker in the promoter region of MELO3C019694 in Harukei-3 genome (Supplementary Data 2). Around this region, two retrotransposon-related protein-coding sequences were located: MELO.jh102710.1 and MELO.jh102711.1. c Tissue-wide gene expression patterns of MELO3C019694 and neighboring retrotransposon-related sequences. Their expression levels were highest in post-harvest ripening fruits (indicated by a yellow box).