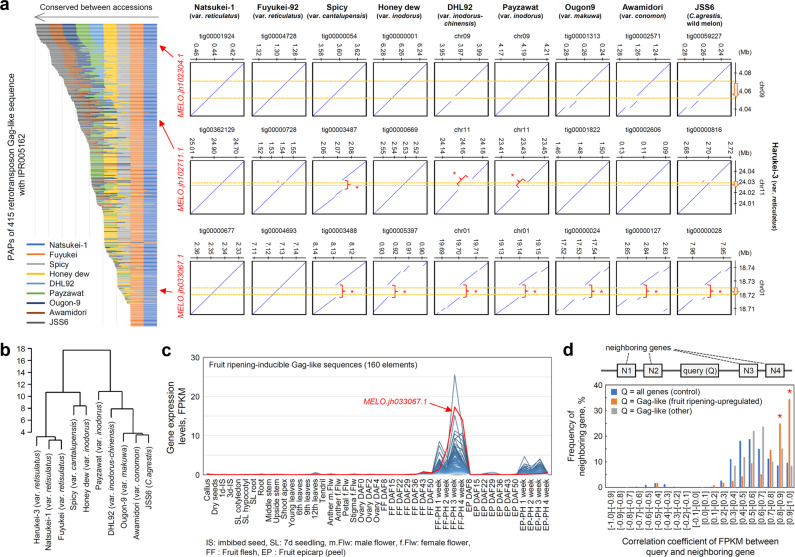
Fig. 5. Presence/absence polymorphism (PAP) and fruit-ripening-inducible expression of retrotransposon Gag-like sequences.
a Assembly-based PAP analysis of 415 retrotransposon Gag-like sequences between melon genomes. PAP was analyzed based on genomic DNA alignment using Harukei-3 as a reference. Except for the DHL92 and Payzawat genomes, genomic contigs were newly assembled based on the Oxford Nanopore technology sequencing data. The left graph shows the summary of the PAP analysis while the right panels show some examples of genomic alignments. In the right panels, genomic regions that are present in Harukei-3 but absent in other melons are indicated by red arrows and parentheses. In each alignment, the positions of Gag-like sequences are shown by yellow dashed lines. b A hierarchical clustering of 10 melon genomes based on the PAP genotyping dataset of Gag-like sequences. The PAP data of 415 Gag-like sequences shown in (a) were subjected to R-hclust analysis. c Fruit ripening-inducible expression of Gag-like sequences. Of the 415 Gag-like sequences found in the Harukei-3 genome, 160 show fruit-ripening-inducible expression. The expression of MELO.jh033067.1 that was present in reticulatus melons but absent in other melons was strongly up-regulated in post-harvest ripening fruits (red arrow). d A histogram showing the correlation of gene expression between the Gag-like sequence and its neighboring genes. Correlation coefficients were calculated based on Fragments Per Kilobase of exon per Million mapped fragments (FPKM; gene expression levels) values from the tissue-wide transcriptome dataset shown in Fig. 3. Three different groups of queries were analyzed; all genes (control), fruit-ripening-inducible Gag-like sequences, and other Gag-like sequences (not up-regulated in ripening fruits, control). In the case of fruit-ripening-inducible Gag-like sequences, co-expression was observed for 59.4% of the neighboring genes (red asterisks).