Abstract
Fungal endophytes can influence production and post-harvest challenges in carrot, though the identity of these microbes as well as factors affecting their composition have not yet been determined, which prevents growers from managing these organisms to improve crop performance. Consequently, we characterized the endophytic mycobiome in the taproots of three carrot genotypes that vary in resistance to two pathogens grown in a trial comparing organic and conventional crop management using Illumina sequencing of the internal transcribed spacer (ITS) gene. A total of 1,480 individual operational taxonomic units (OTUs) were identified. Most were consistent across samples, indicating that they are part of a core mycobiome, though crop management influenced richness and diversity, likely in response to differences in soil properties. There were also differences in individual OTUs among genotypes and the nematode resistant genotype was most responsive to management system indicating that it has greater control over its endophytic mycobiome, which could potentially play a role in resistance. Members of the Ascomycota were most dominant, though the exact function of most taxa remains unclear. Future studies aimed at overcoming difficulties associated with isolating fungal endophytes are needed to identify these microbes at the species level and elucidate their specific functional roles.
Subject terms: Microbiome, Agroecology
Introduction
Carrot (Daucus carota L. subsp. sativus (Hoffm.) Arcang.) is one of the most important vegetable crops in the world, providing a good source of beta-carotene, fiber, Vitamin A and other vitamins and minerals to the human diet1,2. Carrot taproots are often consumed raw, with per person consumption averaging 3.8 kg in 20153. Organic carrot production now accounts for 14% of the U.S. market4, and price premiums average 15%4, representing an opportunity for growers to transition to organic production. However, both organic and conventional carrot growers face many challenges to produce quality crops while protecting the environment. For example, while carrots are considered a nitrogen (N) scavenging crop, a substantial amount of N fertilizers are lost to the environment5,6. Carrots are also subject to attack by many pests and diseases including Alternaria dauci7, and root knot nematodes8, as well as those that contribute to post-harvest storage losses9.
Endophytes, which are now commonly defined as microbes that spend at least part of their life cycle living inside plant tissues10, are one component of the plant microbiome that could help address these challenges. These microbes have been demonstrated to help plants acquire nutrients11–13, withstand abiotic stress14,15, and possibly even enhance the nutritional quality of crops. For example, some endophytes can produce or stimulate production of secondary metabolites16,17, indicating that they could play a role in the nutritional quality and organoleptic properties of plants18. In addition, many endophytic taxa, especially fungi, have been shown to reduce disease caused by pathogenic bacteria, fungi and nematodes19–23, via mechanisms that include competition, antibiosis, parasitism and induction of systemic resistance24. In fact, fungal endophytes could be particularly well suited to act as biocontrol agents, because they occupy the same ecological niche as invading pathogens25. Moreover, they would not need to compete with other soil microbes, which reduces the efficacy of many biocontrol products26.
While fungal endophytes clearly have potential to suppress diseases and improve performance in crops like carrot, the exact functional roles of many of these microbes remain unclear, which prevents their exploitation in agricultural systems. In addition, some fungal endophytes could negatively affect plant and possibly even human health. For example, while endophytes were originally defined as microbes that “can be isolated from surface disinfected plant surfaces” and “do not visibly harm the plant”27, this definition is now widely regarded as problematic because not all endophytes are culturable, and it is not easy to assess phytopathogenicity or distinguish latent pathogens from endophytes14,15. Moreover, some fungal endophytes can act synergistically with pathogens to facilitate infection and/or accelerate disease symptoms19,22,28. Antagonism appears to be the most common life history trait among fungal endophytes, though these relationships can be context dependent for reasons that are still unclear28. In addition, while many fungal endophytes are expected to be mutualists29,30, with both partners benefiting from the relationship, some appear to act as commensals gaining resources without providing any obvious benefits31. Finally, some endophtyic taxa with so-called ‘plant growth promoting properties’, can act as opportunistic pathogens in humans32. Consequently, additional studies are needed to determine how the benefits of mutualistic fungal endophytes can be leveraged, while minimizing the potentially negative effects of others.
Endophytes generally represent a subset of microbes in bulk soil, indicating that plants have some degree of control over which taxa are allowed to enter33–35. Nevertheless, soil is critical in shaping endophyte communities36,37 , since most endophytes are horizontally transmitted19,38. Consequently, crop management practices that alter soil microbial communities are likely to be critical in the composition and functional role of endophytes. For example, in a recent study, we demonstrated that carrot taproots grown in an organic cropping system hosted a greater abundance and diversity of culturable endophytes that could suppress A. dauci than carrots grown in a conventional system39. Another factor that can play a role in shaping plant microbiomes is plant genotype40. Moreover, some studies have demonstrated that microbiomes differ between genotypes that are resistant and susceptible to phytopathogens, indicating that these communities could play a role in these critical plant traits41–43, and this could be the case for carrot. For example, we recently conducted a greenhouse trial using field soil collected from organic and conventional management systems that were expected to be ‘disease suppressive’ and ‘disease conducive’, respectively, based on the results of our previous field trial39. Interestingly, only the nematode resistant genotype (E3999) had greater yield in pots containing the organic soil inoculum than those with the conventional inoculum or a sterile control44. Consequently, we suspect that this genotype might be able to recruit beneficial microbes when they are present in soil to aid in pathogen resistance, and/or provide other growth promoting properties such as better access to nutrients.
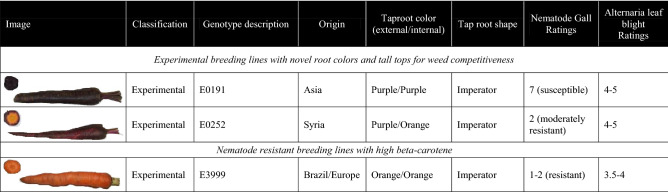
The development of new high-throughput sequencing technologies has made it possible to overcome limitations associated with isolating and culturing endophytic microbes and begin to investigate their potential functional role. Several studies have used these technologies to identify endophytic taxa in model crops such as Arabidopsis and Medicago36,45, as well as major agronomic crops such as maize46,47, however other important crops like carrot have been overlooked. Consequently, the objective of this study was to determine how management system and carrot genotype interact to affect the composition of fungal endophyte communities using culture-independent sequencing technologies. We predicted the following: (1) fungal endophyte communities would be more diverse in taproots grown in the organic system due to greater abundance and diversity of soil microbes; (2) carrot genotypes would host distinct communities due to differences in resistance to pathogens; and (3) the resistant genotype would be most responsive to management system, because the resistance of this genotype is due, at least in part, to its ability to recruit antagonistic fungi and/or prevent colonization of taxa that promote disease severity. To test these hypotheses, we selected three experimental genotypes that vary in resistance to root-knot nematodes and A. dauci (Table 1). The carrots were grown in a long-term trial comparing organic and conventional farming systems, and the composition of fungal endophyte communities in carrot taproots was identified via Illumina sequencing of internal transcribed spacer (ITS) fragments.
Table 1.
Carrot genotypes grown in conventional and organically managed systems at Purdue’s Meigs Farm during summer 2015.
Material and methods
Field trial
Carrot taproots were grown in a long-term crop systems trial comparing organic (ORG) and conventional (CNV) management at Purdue’s Meigs Horticultural Research Farm (lat. 40°17′21″ N. long. 86°53′02″), located approximately 10 miles south of Lafayette, IN during summer 201548. Soil at this site is classified in the Drummer soil series, which typically contain approximately 3.2% organic matter and a neutral pH. The mean annual precipitation at this site is 1,008 mm, and summer temperatures range from 21.1 to 26.7 °C. The crop systems trial was established in 2011 on adjacent tracts of land with uniform topography that had previously been managed using either organic or conventional farming practices since 2001. The crop systems trial was arranged in a split-block design with three replicates for each system given constraints at the site. Within each crop system, four cash crops, carrot, tomato (Solanum lycopersicum), popcorn (Zea mays everta) and soybean (Glycine max), were grown annually and managed using standard practices for each system. This included application of inorganic fertilizers and synthetic pesticides in the conventional system, and inclusion of a winter cover crop and organic fertilizers in the organic system. The winter cover crop planted in the organic system consisted of a custom fall green manure mix containing winter rye (Secale cereale L.), hairy vetch (Vicia villosa), winter pea (Pisum sativum), annual rye (Lolium multiflorum), and timothy grass (Phleum pratense) (Cloverland Seed, Millersburg, OH). Cash crops were rotated in both crop systems annually in the following order: tomato—> carrot—> popcorn—> soybean.
In the carrot plots, fertilizers were applied to both systems to achieve a target rate of 134.5, 180 and 224 kg ha−1 of N, P and K respectively. In the organic plots, this consisted of Re-vita Pro Compost (Ohio Earth Foods, Hartville, OH), applied at a rate of 5,380 kg ha−1 to meet fertility needs, assuming 50% of the nutrients would be available for plant uptake in the year of application. In the conventional plots, diammonium phosphate (18-46-0) and potash (0-0-60) were applied to meet fertility needs. Sub-plots containing 36 carrot genotypes, which represented advanced breeding lines as well as commercial check cultivars, were randomized within each larger carrot plot, for a total of three replicates per crop system. Three of these carrot genotypes (E0191, E0252, E3999) were selected for further analysis of their endophyic mycobiome based on their country of origin, differences in top size and tap root color/shape, and resistance to pathogenic soil nematodes and A. dauci (Table 1). Untreated carrot seeds provided by the USDA-ARS Vegetable Crop Research Unit, Madison, WI, were planted in mid-May. Seeds were planted on raised beds that were 1.8 m apart, in 1 m rows to provide approximately 60 plants m−1 per sub-plot given previously determined germination rates. Seeds were sown to a depth of 1 cm. In the conventionally managed system, a pre-emergent herbicide (Prowl H2O, BASF Corporation) was applied immediately after planting. In the organically managed system, plots were hand weeded as needed. No additional pesticides were applied in either crop management system.
Carrot screening for foliar and soil-borne pathogens
The percentage of infection by foliage pathogens in each plot was quantified using the Horsfall-Barret rating scale49 60 and 110 days after seeding. In brief, the percentage of leaf area showing blight symptoms in each plot was assigned a numerical value from 1 to 12 using the arbitrary Horsfall-Barratt rating scale in which 1 = 0% infection and 12 = 100% infection. At harvest (110 days after seeding), carrots were manually harvested, and the presence of any galls or forking to indicate damage by root knot nematodes, and total number and weight of all taproots, and weight of aboveground foliar in each plot were recorded.
Soil chemical and biological assays
Ten soil cores were randomly collected to a depth of 10 cm in each field rep just prior to carrot seeding in spring. The ten cores within each field rep were pooled and transferred to the laboratory on ice. After thoroughly mixing the cores from each replicate, a subsample of soil was air-dried before shipping to Midwest Labs (Omaha, NE) for a standard soil test according to common methods used in this region50. Briefly, total organic matter was determined using loss of weight on ignition; available P was extracted as Weak Bray (readily available P) and Strong Bray (potentially available P) and analyzed calorimetrically; exchangeable potassium (K), calcium (Ca), and magnesium (Mg) were extracted with neutral ammonium acetate (1 N) and quantified by inductively coupled argon plasma–mass spectrometry detection; and base saturation and cation exchange capacity [mmol ( +)·kg−1] were estimated from the results of exchangeable minerals50. Another subsample was placed in the cooler at 4 °C until being air-dried overnight to conduct assays to estimate microbial activity and active soil carbon. Microbial activity was estimated using the hydrolysis of fluorescein diacetate (FDA) in soil slurries using a method optimized for soil51. Active C was quantified using the permanganate oxidizable carbon (POXC) technique52. Finally, a subsample was lyophilized and stored at − 20, before being shipped overnight on dry ice to WARD lab (Grand Island, NE) for phospholipid fatty acid analysis (PLFA) using methods described in53.
Statistical analysis of soil and plant assays
All soil chemical properties, soil microbial biomass and activity, percent infection of aboveground foliage, and number and weight of carrot roots and shoots were statistically analyzed using the general linear model procedure for ANOVA, and differences among treatment pairs were determined using the student’s t test at a p-value of 0.05, using the SAS JMP software package54. All data were checked for normality, homogeneity of variance and linearity prior to analysis, and were transformed when necessary.
Fungal endophyte DNA extraction, amplification and sequencing
At harvest, two randomly selected carrot taproots representing each genotype selected for the endophytic mycobiome analysis (E0191, E0252, E3999), were collected from each of the field replicates, placed in a cooler on ice and transferred to the lab where they were stored at 4 °C until processing within 48 h. Taproots were collected from healthy plants with no signs of disease or any other plant stress. The taproots were rinsed thoroughly with tap water, then surface disinfected by soaking in 5.25% bleach for 3 min, followed by soaking in 3% peroxide solution for 3 min, and finally washing with sterilized water supplemented with 1% tween55. To confirm surface disinfection of the carrot taproots, 200 µl samples from the last washing solution were plated onto semi-selective media for heterotrophic bacteria (Tryptic Soy Agar), oligotrophic bacteria (R2A), and total fungi (1/5th PDA media)56,57, each with two replicates. The carrot cores were also rolled over the surface of each semi-selective media. The petri plates were incubated at 27 °C or 25 °C and counted after 48 or 72 h, for bacterial and fungal enumeration respectively. Five (15 mm) carrot cylinders were collected from each taproot using a sterilized core borer, and the five cores from each field replicate were pooled for analysis. Carrot core samples were lyophilized (LABCONCO, Kansas City, U.S.A) and stored at − 80 °C until DNA extraction.
Endophyte community DNA was extracted in duplicate from each lyophilized carrot root sample using Qiagen DNeasy Plant Mini Kits (Qiagen, U.S.A) following the manufacturer’s protocol and diluted using 100 μl of elution buffer. The two lab replicates were pooled, and DNA was quantified using a Qubit Fluorometer 2.0 and dsDNA HS Assay Kit (Thermo Fisher Scientific, U.S.A.) and normalized to 1 ng/μl prior to ITS amplification. Fungal endophyte community ITS library construction was carried out in two steps. First, the ITS1 region was amplified using the universal primers ITS1F forward primer 5 ′CTTGGTCATTTAGAGGAAGTAA-3′58 and ITS2 reverse primer 5′-GCTGCGTTCTTCATCGATGC-3′59 modified to contain an adapter region for sequencing on the Illumina MiSeq platform, in triplicate reactions for each sample. Each 25-μl PCR reaction mixture contained 3 μl of DNA template, 0.5 μl (100 mM) of each primer, 12.5 μl GoTaq colorless Master Mix (Promega, Wisconsin, U.S.A) and 8.5 μl of nuclease free water (Promega, Wisconsin, U.S.A.). Each PCR reaction was performed using a Bio-Rad T100 Thermal Cycler (BioRad, California, U.S.A) with the following conditions: initial denaturing using 1 cycle at 95 °C for 2 min, 40 cycles of the following (denaturing step 95 °C for 30 s, annealing step 55 °C for 30 s, and extension step 72 °C for 1 min), and a final extension step of 72 °C for 10 min. Detection of PCR-amplified products was performed with electrophoresis on a 0.7% (wt. /vol.) agarose gel stained with Bullseye DNA Safe Stain (MIDSCI, U.S.A.). A 100 bp ladder (New England bio lab, U.S.A) was also run in parallel to approximate PCR product band sizing. Presence of DNA bands stained with DNA Safe Stain (MIDSCI, U.S.A.) were visualized after exposure of the gel to ultraviolet (UV) light. PCR replicate products of the same samples were pooled and cleaned using Ultraclean PCR Clean-Up Kits (MO BIO, U.S.A) following the manufacturer’s protocol. Cleaned PCR products were subjected to a second PCR reaction, with specific tag encoded primers for each sample. The same thermocycling conditions described above were used, with the exception of 5 amplification cycles instead of 35. Again, all PCR products were confirmed by electrophoresis as described above. Final PCR product concentration was quantified and adjusted using the Qubit Fluorometer 2.0 as described above. Samples were submitted in equimolar concentrations (20 ng) to the Purdue Genomics Facility for sequencing of ITS libraries. A TruSeq DNA LT Sample Prep Kit (Illumina, San Diego, CA) was used to construct paired-end (2 × 250 bp) sequencing libraries. MiSeq Reagent Kit v2 (Illumina, San Diego, CA) was used to perform amplicon sequencing on a MiSeq Desktop Sequencer (San Diego, CA).
After demultiplexing, the reads were quality-filtered, converted to FASTA format using FASTX-toolkit (Version 0.0.14), and concatenated into a single file for use as an input into QIIME (Version 1.9.1)60. The reads with Phred quality score of Q30 were retained for further analysis. Operational taxonomic unit (OTU) picking, taxonomic assignment, and construction of phylogenetic trees were carried out using QIIME’s open-reference OTU picking module using the UCLUST method61. Reads were clustered against a reference fungal database (UNITE 97, 12_11 version) at 97% identity, and reads that failed to hit the reference were subsequently clustered de novo into operational taxonomic units (OTUs). All the samples were taken into account without any subsampling. The suppress_align_and_tree was passed as a parameter because the trees generated from ITS sequences are generally not phylogenetically informative. Only OTUs of fungal origin were considered for further analysis. The QIIME module identify_chimeric_seqs.py that employs the Chimera Slayer algorithm62 was used to screen for chimeric sequences. To report the number of sequences per sample, the QIIME module biom summarize-table was used. To estimate the alpha diversity within the taproots of three carrot genotypes grown under organic and conventional management , the alpha diversity script based on Faith’s phylogenetic diversity index63 was used. The two-sample t-test was used to determine the diversity between genotypes under different soil management. The diversity in the samples was calculated using three different diversity indices: Observed OTUs, Chao-1 Estimator64, and PD_whole_tree65 and sequencing depth was assessed using rarefaction curves.
QIIME’s filter scripts were used to retain OTUs where 25% of the samples in groups being compared have OTUs. Beta diversity estimates were calculated within QIIME using Bray–Curtis distances matrices and results were used to produce principle coordinate analysis (PCoA) plots to visualize differences66. Community differences within all samples of a group as well as between different groups were further assessed using t-tests, while community differences between groups were assessed using QIIME’s compare_categories.py script and ADONIS methods67.
In order to quantify differential abundance for specific OTUs between groups among the different comparisons, the phyloseq software package, implemented in Bioconductor, was used to provide a platform for statistical analysis and figure generation in R For each comparison, p-values were adjusted for the false-discovery rate (FDR) and OTUs with adjusted p-values below 0.2 were considered significant and were used to generate ggplot2 summary plots. Finally, to determine which fungal OTUs best characterized taproot endophyte communities as a function of management system, carrot genotype, and the interaction of these two factors, we used an indicator species analysis in the labdsv package in R68. Indicator species values are based on how specific and widespread an OTU is within a particular subgroup and are independent of the relative abundance of other fungal taxa in carrot taproots69.
Results
Impact of management system on soil properties, disease severity and yield
Soil pH, total and active organic matter, calcium and percent calcium on cation exchange sites (CEC) were significantly greater in the organic system, while percent hydrogen on CEC sites was significantly greater in the conventional system (Table 2). Many components of the microbial biomass including total microbes, total bacteria, gram positive and negative bacteria, actinomycetes, and total fungi were greater in the organic system (Fig. 1). The severity of leaf blight caused by foliar pathogens was high during summer 2015 in the carrot plots as all carrot genotypes had between 75–90% infections just prior to harvest (119 days after seeding), but there were no significant differences between carrot genotypes or management system (Table 3). Pathogens isolated from carrot foliage in both of these cropping systems have previously been identified as A. dauci, Cercospora carotae and Xanthomonas campestris70 (duToit, personal communication). There were no visible symptoms of nematode infection in any of the three carrot genotypes evaluated in this study regardless of the susceptibility or resistance to nematodes (Table 3). Total shoot and root weight in genotype E0252 was greater in the conventional than organic system, and the shoot and root weight of E0252 was greater than E3999 (Table 3).
Table 2.
Soil chemical properties, active organic matter and microbial activity in carrot field managed using organic and conventional farm practices just prior to planting in summer 2015 at Purdue’s Meigs Horticulture Research Farm.
| Crop system | %OM | P-weak bray | P-strong bray | K | Mg | Ca | pH | CEC | %K | %Mg | %Ca | %H | POXC | FDA |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ppm | Percent base saturation | mg POXC/kg soil | ug FDA/g soil/h | |||||||||||
| Organic | 3.1 az | 34.3 | 67.7 | 230.0 | 426.3 | 2,790 a | 6.7 a | 19.2 | 3.1 | 18.3 | 72.8 a | 4.5 b | 395.2 a | 0.162 |
| Conventional | 2.2 b | 70.7 | 81 | 256.3 | 335.7 | 1991 b | 6.0 b | 16.0 | 4.1 | 17.5 | 62.6 b | 15.7 a | 294.9 b | 0.122 |
zDifferent letters within a column represent significant difference as determined by Tukey’s honestly significant difference test (P < 0.05).
Figure 1.
Microbial biomass estimated using soil phospholipid fatty acid analysis (PLFA) in soil collected from carrot plots grown using organic and conventional management at Purdue’s Meigs Farm during summer 2015. zDifferent letters within a column represent significant difference as determined by Tukey’s honestly significant difference test (P < 0.05).
Table 3.
Carrot biomass, percentage of damage by foliar pathogens and nematode diseases severity in organic and conventional field trials during summer 2015.
| Management system | Carrot genotype | % Damage by foliar pathogens | # of plants | Nematode rating | Plant biomass at harvest (g) | ||||
|---|---|---|---|---|---|---|---|---|---|
| 60 day | 119 day | Shoots | Roots | ||||||
| Organic | E0191 | 0.0 | 71.7 | 8.3 | 0.0 | 0.19 | A | 0.57 | A |
| Conventional | E0191 | 3.3 | 83.3 | 5.3 | 0.0 | 0.23 | 0.59 | ||
| Organic | E0252 | 5.0 | 90.0 | 3.7 | 0.0 | 0.05 b | A | 0.14 b | AB |
| Conventional | E0252 | 0.0 | 66.7 | 9.7 | 0.0 | 0.22 a | 0.64 a | ||
| Organic | E3999 | 23.3 | 91.7 | 5.7 | 0.0 | 0.02 | B | 0.12 | B |
| Conventional | E3999 | 25.0 | 83.3 | 7.3 | 0.0 | 0.04 | 0.27 | ||
zDifferent letters within a column represent significant difference as determined by Tukey’s honestly significant difference test (P < 0.05).
Abundance and quality of fungal endophyte sequences
After quality filtering, adapter trimming, and merging of Illumina reads, approximately 3,793,627 high-quality sequences were obtained and used as input for analysis and comparison of fungal endophyte communities. Sequences clustered into 1,480 different fungal operational taxonomic units (OTUs) when grouped at the 97% genetic similarly level (Table S1). Rarefaction curves (Fig. S1) indicated that only 38.5% of fungal endophyte diversity present in carrot taproots was recovered by this surveying effort.
Assignment of OTUs to fungal taxa
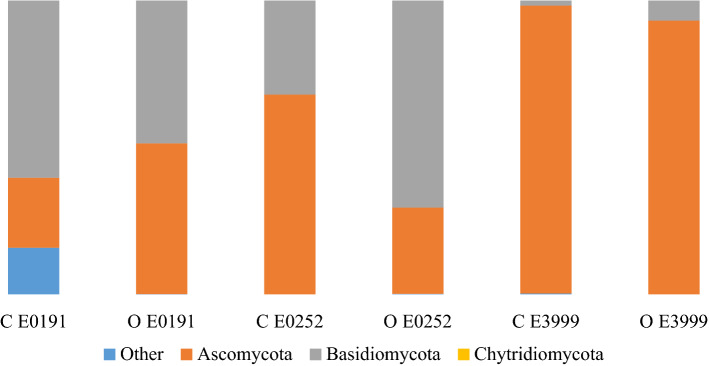
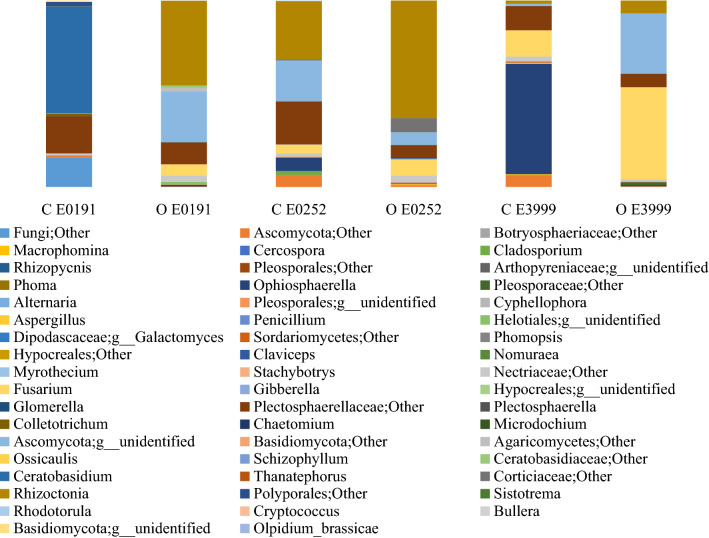
Carrot taproots were dominated by microbes in the Ascomycota phyla (73.9%) (Fig. 2). Other abundant phyla belonged to the Basidiomycota (24.8%) and Chytridiomycota (< 1%) (Fig. 2). At the level of genera, Rhizoctonia and Fusarium were predominant, representing 19% and 13% of all endophytes identified (Fig. 3). Other taxa observed across all samples included Ophiosphaerella (5.4%), Ceratobasidium (3.6%), Colletotrichum and Gibberella (each at 0.4%), Cladosporium (0.3%), Aspergillus (0.2%), and Cyphellophora, Thanatephorus, Alternaria and Plectosphaerella (all at 0.1%). Finally, Cercospora, Rhizopycnis and Phoma were among twenty other genera observed with less than 0.1% relative abundance.
Figure 2.
Relative abundance of fungal endophytes by phyla in the taproots of three carrot genotypes grown under conventional (C) and organic (O) management.
Figure 3.
Relative abundance of fungal endophytes by genera in the taproots of three carrot genotypes grown under conventional (C) and organic (O) management.
Effect of crop management system on fungal endophytes
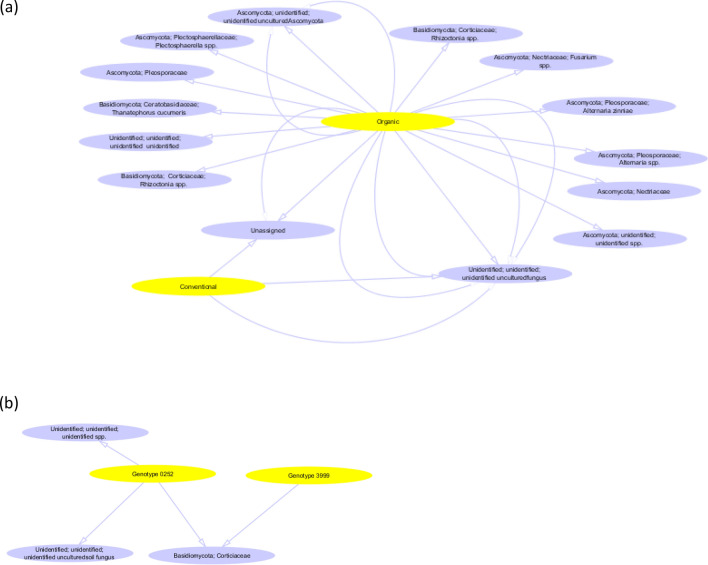
Fungal endophyte richness (Table 4a) and beta diversity (Table 4b) were significantly greater in the organic management systems, but alpha diversity was not (Table 4a). Of the 1,480 individual fungal endophyte OTUs identified, 98.3% were not significantly different in relative abundance or frequency with respect to management system (Fig. 4a & Table S2). However, individual OTUs representing Ascomycota, Basidiomycota and Chytridiomycota phyla, which comprised 1.6% of all fungal taxa observed in the study, were specifically associated with one management practice (Fig. 4a and Table S2). Of these, 87.5% were significantly associated with organic management, while only one unidentified, unassigned and uncultured fungal genus was significantly associated with conventional management. At the level of genera, the indicator species analysis indicated that genera belonging to Alternaria, Fusarium Plectosphaerella, Rhizoctonia and Thanatabasidium were uniquely correlated with organic management, while only one unidentified and one unassigned species was uniquely correlated with conventional (Fig. 4a).
Table 4.
(a) Influence of crop management systems, carrot genotype and their interactions on fungal endophyte richness and alpha diversity within the taproots of three carrot genotypes grown under organic and conventional management. (b) Influence of crop management systems, carrot genotype and their interactions on fungal endophyte beta diversity within the taproots of three carrot genotypes grown under organic and conventional management.
| (a) Comparison | Richness | Diversity |
|---|---|---|
| p-value | p-value | |
| Management system | 0.019 | 0.354 |
| Carrot genotype | 0.778 | 0.205 |
| Management system + carrot genotype | 0.284 | 0.524 |
| Management system + E0191 | 0.275 | 0.513 |
| Management system + E0252 | 0.513 | 0.827 |
| Management system + E3999 | 0.050 | 0.275 |
| (b) Comparison | Bray–Curtis | |
|---|---|---|
| p-value | ||
| Management system | 0.030 | |
| E0191 vs. E0252 | 0.743 | |
| E0191 vs. E3999 | 0.720 | |
| E0252 vs. E3999 | 0.667 | |
| Conventional E0191 vs. conventional E0252 | 0.801 | |
| Conventional E0191 vs. conventional E0252 | 0.800 | |
| Conventional E0252 vs. Conventional E3999 | 0.901 | |
| Organic E0191 vs. Organic E0252 | 0.801 | |
| Organic E0191 vs. organic E3999 | 0.801 | |
| Organic E0252 vs. organic E3999 | 0.900 | |
| Conventional E0191 vs. organic E0191 | 0.201 | |
| Conventional E0252 vs. organic E0252 | 0.900 | |
| Conventional E3999 vs. organic E3999 | 0.101 |
Figure 4.
Indicator species analysis identifying individual fungal OTUs in carrot taproots; (a) Fungal OTUs unique to management system, (b) Fungal OTUs unique to carrot genotype.
Effect of carrot genotype on fungal endophytes
Carrot genotype did not affect fungal richness, alpha (Table 4a), or beta diversity (Table 4b) when genotypes were compared across management systems. However, several individual fungal genera including unidentified and non-assigned genera, Cladosporium, Thanatephorus, Rhizoctonia, Ceratobasidium, Aspergillus, Cyphellophora and Ophiosphaerella differed among genotypes (Fig. S2). Specifically, there was a greater abundance of Cladosporium, Thanatephorus, Rhizoctonia, Ceratobasidium and Aspergillus in E0191 when compared with E3999, whereas the opposite occurred with Cyphellophora and Ophiosphaerella genera. E0191 also had a greater abundance of Aspergillus and Ceratobasidium than E0252. In contrast, only a few unidentified and non-assigned genera were more abundant in E0252 than E3999 (Fig. S2). The indicator species analysis indicated that only three out of the 1,480 fungal taxa were correlated with an individual carrot genotype (Fig. 4b and Table S3). This included one uncultured fungus and one unidentified fungus that were uniquely associated with E0252, and one fungal taxon related to the Corticiaceae family, that was correlated with E0252 and E3999.
Interactions between carrot genotype and management system on fungal endophytes
Only genotype E3999 had differences in taxonomic richness of fungal endophytes (P < 0.05) (Table 4a), and marginal differences in beta diversity (P < 0.10) (Table 4b) when grown in the two contrasting production systems. There were differences in the abundance of individual fungal endophyte OTUs in E0191 and E3999 grown in the two management systems (Fig. 3; Fig. S2). Specifically, within E0191, the relative abundance of Aspergillus, Ophiosphaerella, Rhizoctonia, Thanatephorus and Fusarium were significantly greater in carrots grown in the organic system whereas the opposite was found with Colletotrichum and Ceratobasidium. Within E3999, seven fungi including an uncultured Ascomycota, an uncultured fungus, and some non-assigned taxa were significantly greater when grown under organic compared to conventional management, while the opposite was found for one uncultured fungus and one non-assigned fungus. In contrast, no differences in individual genera were detected in E0252 when grown in the organic compared to conventional management system (Fig. S3).
When comparing differences among genotypes within each individual management system, there were differences in the abundance of some genera (Fig. S2). Specifically, under conventional management, a greater relative abundance of Ophiosphaerella and Cladosporium genera were present in E0252 than E0191, whereas the opposite was found for Ceratobasidium. Ophiosphaerella was more abundant in E3999 than either E0191 or E0252, along with a few other unidentified and non-assigned genera (Fig. S2). Under organic management, Aspergillus and Rhizoctonia were more abundant in E0191 than E3999, one unidentified genus had greater abundance in E0191 than E0252, and one non-assigned genus had greater abundance in E0252 than E3999 (Fig. S2).
Discussion
Results of this study confirm earlier reports9,39 indicating that carrot taproots are colonized by a diverse assortment of fungal endophytes, with the majority belonging to the Ascomycota phyla (Fig. 2). Members of the Ascomycota are common as endophytes in the roots of a wide variety of plant species in ecosystems ranging from the arctic tundra, to tropical forests and croplands9,71–75. In a recent review of all eukaryotic ITS sequences available, the Ascomycota represented 30% of all fungal endophytes in plant roots identified to date14. It is unclear why these fungi are so predominant in plant roots and especially in carrot, though it could have something to do with their close relationship with many pathogens in the same phyla14,76. To enter and survive inside plants as endophytes or pathogens, microbes must possess plant-degrading enzymes, and/or be able to silence plant defense pathways36,77. Over the course of evolution, there are many examples of transitions between endophytic and pathogenic life history traits among the Ascomycota78. Consequently, endophytism could remain a viable life history strategy if members of the phyla act as ecological opportunists to form pathogenic relationships when environmental conditions make this strategy better for their long-term survival78. This could also help explain why many members of this phyla act as pathogens in one plant species and endophytes in another14, especially if they are obligate microbes that cannot survive or reproduce in soil.
Other prominent fungal phyla in carrot taproots included members of the Basidiomycota (Fig. 2). Fungal taxa within the Basidiomycota also include mutualists and commensals as well as pathogens14,19, so their predominance in carrot taproots could also be related to their ability to transition between endophytism and parasitism. Surprisingly, we did not observe any fungi from the Glomeromycota, despite the fact that they are generally the most abundant fungal endophytic phylum in plant root surveys14. Members of the Glomeromycota form arbuscular mycorrhizas, which are well known for their potential to help plants, including carrot, obtain nutrients and withstand biotic and abiotic stress79,80. This could be due to the fact that the primer sets we used are not ideal for amplifying this fungal phylum58,81,82, as well as that our samples were from carrot taproots rather than fine roots where mycorrhizal fungi are generally more common9,83. Unfortunately, many of the OTUs obtained in this study were characterized as either unidentified or unassigned, highlighting the challenges associated with the lack of informative sequences in existing fungal databases82,84.
As 98.3% of the fungal taxa identified in this study did not differ between the two management systems (Table S2; Fig. S2), these taxa likely represent a ‘core mycobiome’ in carrot taproots. A plants core microbiome represents a set of microbial taxa that are systematically associated with a given host plant85. In many cases, these core microbiomes appear to remain relatively stable over the course of evolution and domestication12,36,86, at least with respect to broad taxonomic groups14. Nevertheless, soil can influence the composition of endophytes34,35,37, especially at finer taxonomic scales. Results of our study provide further support for this phenomenon, by demonstrating that the diversity of fungal endophytes in carrot taproots is dependent on crop management systems that differ in soil chemical and biological properties (Tables 2 and Fig. 1). Other studies have also provided evidence that differences in soil characteristics induced by management practices are a strong driver of endophyte composition55,87,88. In particular, management practices commonly used in organic and conventional farming systems are well known for their potential to alter many soil properties89. For example, organic farmers commonly plant cover crops and apply organic fertility amendments, which increase soil organic matter and serve as the primary food and energy source for soil microbes89–91. Consequently, as the soil in the organic management system in this study had more active organic matter and a greater abundance of several types of soil microbial biomass including fungi (Table 2 and Fig. 1), it is not surprising that endophytes were more diverse in taproots grown in this system (Fig. S2). Others have suggested that differences in fungicide applications between organic and conventional systems could also affect endophyte composition87,92, however, this is not likely to be the case in this study, as we did not apply any fungicides in either system.
While clarifying the specific functional roles of fungal endophytes in carrot taproots will require additional studies using taxa that have been isolated and cultured, it is possible to begin to speculate about their potential functional roles given results of this sequencing effort. Several individual OTUs were uniquely associated with the organic system (Fig. 4a). While fungi associated with these genera have been implicated as pathogens in some crops, they have also been isolated from healthy plant tissues in other species and demonstrated to provide benefits14,19, indicating that they might not necessarily act as pathogens in carrot. For example, Plectosporella species have been isolated from healthy soybean93, vegetable94, and quinoa95 roots. While some Plectosporella isolates caused disease symptoms when inoculated onto lettuce, others increased plant growth94. In another study, an endophytic Plectosporella isolated from carrot taproots failed to produce any disease symptoms when re-inoculated onto new carrot plants96, indicating that these taxa may not act as pathogens in carrot and instead could provide benefits.
It is possible that some of the taxa that were more abundant in the organic taproots such as Rhizoctonia, are latent pathogens and/or could contribute to diseases caused by other pathogens. However, we do not expect that this was the case here. In our previous study isolating culturable endophytes from carrot taproots grown in the same organic and conventional fields, foliar disease incidence was lower in the organic system in two of the genotypes evaluated in this study (E0191 and E0252)39. Moreover, soils in the organic system had greater microbial biomass and activity, and endophytic isolates collected from roots grown in the organic system had greater antagonistic activity against A. dauci. Consequently, because several soil biological properties were also greater in the organic system in this study (Table 2 and Fig. 1), we expect that microbes in these soils could have been more suppressive against pathogens, and/or had other plant growth promoting properties. Other studies have demonstrated that soils in organic farming systems can be more disease suppressive than their conventional counterparts97,98, and microbes isolated from the rhizosphere of plants grown in organic systems have greater potential to suppress diseases99. Endophytesisolated from vegetables grown under organic management have also been shown to be more abundant and diverse, and have greater growth promoting properties than those grown in conventional systems88. Finally, the fungal endophytes in this study were collected from healthy plants in a year where foliar disease pressure was very high (Table 3), thus we expect that they were not pathogens and instead could have played a role in helping carrots resist diseases, though future studies are needed to verify this hypothesis.
Like pathogens, plants are able to sense and respond to the presence of endophytic microbes, acting as ‘gate keepers’, to exclude or permit different taxa from entering and persisting in plant roots77,100. Consequently, it is not surprising that plant genotype can also play a smaller, yet significant role in shaping plant microbiomes40,101, and carrot is not an exception (Figs. S2, S3). Over the course of evolution and breeding, plants experience different selection pressures which could influence whether the presence of endophytic taxa are maintained15. For example, fungal diversity in plant genotypes selected in modern agricultural systems has been reported to be lower than in wild ancestors, in a phenomenon referred to as “domestication syndrome”102. In contrast, it is also possible that selection for traits such as disease resistance could have inadvertently selected for microbes that aid in plant resistance. For example, targeted breeding efforts have resulted in the development of carrot genotypes that are highly resistant to root knot nematodes103. Mechanisms appear to include: (1) differences in chemical cues attracting nematodes to roots, and the ability of nematodes to (2) penetrate the epidermis, (3) migrate through the root surface to establish a feeding site in the vascular parenchyma, (4) develop root galls, and (5) reproduce103. While some of this resistance is likely regulated by specific R genes, such as those that mediate a hypersensitive response at the root surface when pathogens attempt to enter host tissue, other components could be mediated, at least in part, by endophytes. For example, while host genes for resistance in Populus represent the strongest and first line of defense against pests, antagonism by fungal endophytes represents an important second line of defense37. Interestingly, resistance to root knot nematodes in carrot appears to be mediated post-infection103, thus it is plausible that fungal endophytes could play a role in preventing nematodes from migrating, forming galls and/or reproducing. The two genotypes that differed most in this study with respect to differences among individual OTUs were E0191 and E3999 (Fig. S2, 3), which are susceptible and resistant, respectively, to pathogenic nematodes. Previous studies have demonstrated that fungal endophytes can suppress disease caused by pathogen nematodes23, therefore it is possible that differences in these endophyte communities could play a role in preventing, or facilitating, the infection and severity of pathogenic nematodes in these genotypes. However, they also differ in taproot color (Table 1), and E0191 had significantly greater yield than E3999 (Table 3), so it is also possible that these factors could have contributed to the differences observed in this trial.
Several individual OTUs were significantly greater in the susceptible (E0191) than resistant (E3999) genotype (Fig. S2). Isolates of both Rhizoctonia and Ceratobasidium have been shown to cause disease or disease like symptoms in carrots104,105, though Ceratobasidium has also been reported to act as a mycorrhiza in orchids106, and suppress diseases in rice107 and cacoa108,109. Cladosporium is a pathogen in spinach110, though these taxa can also enhance plant growth in soybean111. Members of the Aspergillus genus have been demonstrated to increase growth and reduce soft rot in carrot plants112. Aspergillus taxa can also produce bioactive products active against many phyto as well as human pathogens113,114, indicating that they could enhance plant as well as human health. However, Aspergillus spp. can also cause human health problems and contribute to reductions in post-harvest quality in carrots115, so isolates of this particular genus would need to be carefully tested before they could be considered for use as inoculants to improve carrot performance32. Three individual OTUs were enriched in E3999 relative to E0191 (Fig. S2). Ophiosphaerella spp. are well known for their potential to act as a pathogen in bermudagrass116, and Cyphellophora endophytes are suspected to play a role in facilitating apple diseases117. However, Ophiosphaerella spp. can solubilize calcium, aluminum and iron phosphates118, indicating that they could play important roles in plant nutrition. Endophytic isolates of Cyphellophora were isolated from plants grown on heavily contaminated mine tailings, indicating that they could play a role in helping plants tolerate abiotic stress100. Members of the Corticaceae are often reported as endophytes in woody plants such as Populus101, though their potential functional role remains unclear. Clearly, there is still much work to do to decipher the actual roles of these fungal taxa in carrot taproots, though now that these taxa have been identified using NGS sequencing, it will be possible to design future studies to isolate these taxa and elucidate their specific role.
We predicted that E3999 would be most responsive to differences in soil microbial communities induced by the management systems evaluated in this trial, because of its disease resistance and the fact that we previously noted increased growth in this genotype in the presence of soil inoculum from the organic system in a controlled trial44. Interestingly, the results of our sequencing efforts support this hypothesis, as E3999 was the only genotype that differed in richness between the two management systems (Table 4a), and there were marginal differences in beta diversity (Table 4b). As described above, organic farming systems can host microbes that promote plant growth and have greater disease suppressive activity than their conventional counterparts39,88,97–99. Consequently, we suspect that there could have been greater populations of fungi with suppressive and/or plant growth promoting activity available in the organic system that could have been recruited by E3999 to help this genotype fight pathogens or improve its growth. Alternatively, it is possible that at least part of the resistant activity of this genotype is due to its ability to restrict entry by endophytic microbes that do not directly cause disease but promote the colonization, survival or virulence of pathogens as part of a pathobiome119. Future studies testing these and other hypotheses are needed to determine the extent to which endophytes can mediate disease dynamics.
We also observed differences in individual OTUs in E0191 when grown under the two cropping systems (Fig S3). Since this carrot genotype lacks genetic resistance, it could theoretically host certain taxa as part of its primary form of defense. Several OTUs were greater in E0191 taproots grown in the organic system (Fig. S3). As described above, Aspergillus isolates can benefit carrots by suppressing soft rot and increasing plant growth112, and endophytic isolates of Ophiospharella can help plants acquire nutrients118. While Fusarium can act as a pathogen in carrot104, many isolates of Fusarium can suppress pathogens including pathogenic Fusarium species. For example, Fusarium endophytes can suppress F. oxysporum pathogens in tomato, and Ustilago maydis pathogens in maize120,121. Two OTUs were significantly greater in E0191 taproots grown in the conventional system (Fig. S3). As described above, Ceratobasidium can act as a pathogen in carrot105, though endophytic isolates of this genera can also help plants acquire nutrients and fight pathogens107–109. Colleotrichum has been noted to act as a carrot pathogen104,122, indicating that this endophyte could make this genotype more susceptible to other diseases. However, endophytic members of the Colletotrichum genus have also been demonstrated to produce bioactive metabolites that work against a number of crop pathogens123,124, and help Arabidopsis plants obtain phosphorous125.
Finally, the one carrot genotype that did differ in yield between the two management systems in this study (E0252) (Table 3), was also the one genotype that showed no difference in endophyte communities between the management systems. This indicates that other factors, such as greater availability of soil phosphorous, or lower pH between the two systems (Table 2), might have been responsible for the greater productivity of this genotype in the conventional system (Table 3). These results also indicate that this particular genotype could be more discriminative in comparison to other genotypes, with respect to permitting colonization of different endophytes s present in field soil, providing further support for genetic controls on endophyte mycobiomes.
Conclusions
Carrot taproots host a diverse assortment of fungal endophytes that appear to be part of a core mycobiome unique to carrot. Nevertheless, crop management practices and genotype play a smaller, yet significant role in shaping these communities indicating that it might someday be possible to leverage these communities to enhance crop performance. Our study is only based on one crop season, so it is possible that these communities could change over time, although it was noted that most fungal endophytes in carrot taproots were consistent across years9 and we expect the same here. Many of the fungi identified in this trial could positively or negatively affect diseases, so difficulties in isolating fungal endophytes must be overcome so researchers can determine their specific functional roles.
Supplementary information
Acknowledgements
This project is funded by NIFA-OREI program through grant #2016-51300-25721, as well as NIFA-Hatch project #’s 1007553 and 1015999. We would like to express our appreciation to Dr. Catherine Aime in reviews of these studies as part of the doctoral dissertation of Dr. Sahar Abdelrazek and Tristand Tucker in supporting studies conducted in the field.
Author contributions
All authors made substantial contributions to this manuscript. Lead author, Dr. S.A., conducted the experiments to collect the data, interpreted the data and wrote the first draft of the manuscript during her PhD studies. Drs. S.C. and J.T. are bioformaticists who conducted the bioinformatic analyses, made figures, wrote the methods and helped Sahar with interpretation of the results. Dr. P.S. and Ms. M.C. are carrot breeders who helped identify the best carrot genotypes to use in the trial, helped design the field trial where the carrots were collected from, helped to obtain federal funding to support these studies, and helped with interpretation of the results. Dr. T.M. helped design the study and provided guidance to Sahar when she was working on the laboratory components of the trial. Drs. Choudhari, Thimmapuram, Simon, Mengiste and Ms. Colley, also reviewed the final draft of the manuscript to offer further suggestions for improvement, and approved its submission. Finally, the corresponding author Dr. L.H., contributed to all parts of the research and manuscript. She was Dr. Abdelrazek’s advisor during her PhD program, who obtained funding to conduct the study, hired Sahar to conduct the work, took the lead in designing the study, and worked with Sahar to interpret the results and improve the initial draft of the manuscript in preparation for submitting for publication. All authors have agreed to be personally accountable for the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-70683-x.
References
- 1.Rubatzky, V.E., C.F. Quiros, and P.W. Simon, Carrots and related vegetable Umbelliferae. 1999: CABI publishing.
- 2.Ahmad T, et al. Phytochemicals in Daucus carota and their health benefits. Foods. 2019;8(9):424. doi: 10.3390/foods8090424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wells HF, Bond JK, Thornsbury S. Vegetables and pulses outlook. Change. 2016;2015:16. [Google Scholar]
- 4.Carlson, A., Investigating retail price premiums for organic foods. Amber Waves, May, US Department of Agriculture, Economic Research Service, Washington, DC, 2016
- 5.Westerveld SM, McKeown AW, McDonald MR. Seasonal nitrogen partitioning and nitrogen uptake of carrots as affected by nitrogen application in a mineral and an organic soil. HortScience. 2006;41(5):1332–1338. [Google Scholar]
- 6.Thorup-Kristensen K. Root growth and nitrogen uptake of carrot, early cabbage, onion and lettuce following a range of green manures. Soil Use Manag. 2006;22(1):29–38. [Google Scholar]
- 7.Dugdale L, et al. Disease response of carrot and carrot somaclones to Alternaria dauci. Plant. Pathol. 2000;49(1):57–67. [Google Scholar]
- 8.Parsons J, et al. Meloidogyne incognita nematode resistance QTL in carrot. Mol. Breed. 2015;35(5):114. [Google Scholar]
- 9.Louarn S, et al. Proteomic changes and endophytic micromycota during storage of organically and conventionally grown carrots. Postharvest Biol. Technol. 2013;76:26–33. [Google Scholar]
- 10.Strobel G. The emergence of endophytic microbes and their biological promise. J. Fungi. 2018;4(2):57. doi: 10.3390/jof4020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mandyam K, Jumpponen A. Seeking the elusive function of the root-colonising dark septate endophytic fungi. Stud. Mycol. 2005;53:173–189. [Google Scholar]
- 12.Johnston-Monje D, Raizada MN. Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS ONE. 2011;6(6):e20396. doi: 10.1371/journal.pone.0020396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newsham KK. A meta-analysis of plant responses to dark septate root endophytes. New Phytol. 2011;190(3):783–793. doi: 10.1111/j.1469-8137.2010.03611.x. [DOI] [PubMed] [Google Scholar]
- 14.Hardoim PR, et al. The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015;79(3):293–320. doi: 10.1128/MMBR.00050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lugtenberg BJ, Caradus JR, Johnson LJ. Fungal endophytes for sustainable crop production. FEMS Microbiol. Ecol. 2016;92:12. doi: 10.1093/femsec/fiw194. [DOI] [PubMed] [Google Scholar]
- 16.Kusari S, Hertweck C, Spiteller M. Chemical ecology of endophytic fungi: Origins of secondary metabolites. Chem. Biol. 2012;19(7):792–798. doi: 10.1016/j.chembiol.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 17.Huang Y-H. Comparison of rhizosphere and endophytic microbial communities of Chinese leek through high-throughput 16S rRNA gene Illumina sequencing. J. Integr. Agric. 2018;17(2):359–367. [Google Scholar]
- 18.Rodríguez P, et al. Are endophytic microorganisms involved in the stereoselective reduction of ketones by Daucus carota root? J. Mol. Catal. B Enzym. 2007;49(1–4):8–11. [Google Scholar]
- 19.Rodriguez R, et al. Fungal endophytes: diversity and functional roles. New Phytol. 2009;182(2):314–330. doi: 10.1111/j.1469-8137.2009.02773.x. [DOI] [PubMed] [Google Scholar]
- 20.Gómez-Lama Cabanás C, et al. The biocontrol endophytic bacterium Pseudomonas fluorescens PICF7 induces systemic defense responses in aerial tissues upon colonization of olive roots. Front. Microbiol. 2014;5:427. doi: 10.3389/fmicb.2014.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Busby PE, Ridout M, Newcombe G. Fungal endophytes: Modifiers of plant disease. Plant Mol. Biol. 2016;90(6):645–655. doi: 10.1007/s11103-015-0412-0. [DOI] [PubMed] [Google Scholar]
- 22.Brader G, et al. Ecology and genomic insights into plant-pathogenic and plant-nonpathogenic endophytes. Annu. Rev. Phytopathol. 2017;55:61–83. doi: 10.1146/annurev-phyto-080516-035641. [DOI] [PubMed] [Google Scholar]
- 23.Schouten A. Mechanisms involved in nematode control by endophytic fungi. Annu. Rev. Phytopathol. 2016;54:121–142. doi: 10.1146/annurev-phyto-080615-100114. [DOI] [PubMed] [Google Scholar]
- 24.Latz MA, et al. Endophytic fungi as biocontrol agents: elucidating mechanisms in disease suppression. Plant Ecol. Divers. 2018;11(5–6):555–567. [Google Scholar]
- 25.Rabiey, M., et al., Endophytes vs tree pathogens and pests: can they be used as biological control agents to improve tree health? Eur. J. Plant Pathol. 2019: p. 1–19.
- 26.Cook RJ. Advances in plant health management in the twentieth century. Annu. Rev. Phytopathol. 2000;38(1):95–116. doi: 10.1146/annurev.phyto.38.1.95. [DOI] [PubMed] [Google Scholar]
- 27.Hallmann J, et al. Bacterial endophytes in agricultural crops. Can. J. Microbiol. 1997;43(10):895–914. [Google Scholar]
- 28.Busby PE, Ridout M, Newcombe G. Fungal endophytes: modifiers of plant disease. Plant Mol. Biol. 2016;90(6):645–655. doi: 10.1007/s11103-015-0412-0. [DOI] [PubMed] [Google Scholar]
- 29.Card S, et al. Deciphering endophyte behaviour: the link between endophyte biology and efficacious biological control agents. FEMS Microbiol. Ecol. 2016;92:8. doi: 10.1093/femsec/fiw114. [DOI] [PubMed] [Google Scholar]
- 30.Card SD, et al. Beneficial endophytic microorganisms of Brassica–A review. Biol. Control. 2015;90:102–112. [Google Scholar]
- 31.May, G., Here come the commensals. Am. J. Bot. 2016. 103. [DOI] [PubMed]
- 32.Hoagland L, et al. Foodborne pathogens in horticultural production systems: Ecology and mitigation. Sci. Hortic. 2018;236:192–206. [Google Scholar]
- 33.Knief C, et al. Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J. 2012;6(7):1378. doi: 10.1038/ismej.2011.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu H, et al. Inner plant values: Diversity, colonization and benefits from endophytic bacteria. Front. Microbiol. 2017;8:2552. doi: 10.3389/fmicb.2017.02552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gottel NR, et al. Distinct microbial communities within the endosphere and rhizosphere of Populus deltoides roots across contrasting soil types. Appl. Environ. Microbiol. 2011;77(17):5934–5944. doi: 10.1128/AEM.05255-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lundberg DS, et al. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488(7409):86. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Philippot L, et al. Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013;11(11):789–799. doi: 10.1038/nrmicro3109. [DOI] [PubMed] [Google Scholar]
- 38.Oono R, et al. Genetic variation in horizontally transmitted fungal endophytes of pine needles reveals population structure in cryptic species. Am. J. Bot. 2014;101(8):1362–1374. doi: 10.3732/ajb.1400141. [DOI] [PubMed] [Google Scholar]
- 39.Abdelrazek, S., Carrot Endophytes: Diversity, Ecology and Function. 2019, Purdue University Graduate School.
- 40.Hoagland L, et al. Key traits and promising germplasm for an organic participatory tomato breeding program in the US midwest. HortScience. 2015;50(9):1301–1308. [Google Scholar]
- 41.Yao H, Wu F. Soil microbial community structure in cucumber rhizosphere of different resistance cultivars to fusarium wilt. FEMS Microbiol. Ecol. 2010;72(3):456–463. doi: 10.1111/j.1574-6941.2010.00859.x. [DOI] [PubMed] [Google Scholar]
- 42.Kwak Y-S, et al. Saccharomyces cerevisiae genome-wide mutant screen for sensitivity to 2, 4-diacetylphloroglucinol, an antibiotic produced by Pseudomonas fluorescens. Appl. Environ. Microbiol. 2011;77(5):1770–1776. doi: 10.1128/AEM.02151-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Upreti R, Thomas P. Root-associated bacterial endophytes from Ralstonia solanacearum resistant and susceptible tomato cultivars and their pathogen antagonistic effects. Front. Microbiol. 2015;6:255. doi: 10.3389/fmicb.2015.00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin R, et al. Unexpected diversity of basidiomycetous endophytes in sapwood and leaves of Hevea. Mycologia. 2015;107(2):284–297. doi: 10.3852/14-206. [DOI] [PubMed] [Google Scholar]
- 45.Bulgarelli D, et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature. 2012;488(7409):91. doi: 10.1038/nature11336. [DOI] [PubMed] [Google Scholar]
- 46.da Silva DAF, et al. Endophytic microbial community in two transgenic maize genotypes and in their near-isogenic non-transgenic maize genotype. BMC Microbiol. 2014;14(1):332. doi: 10.1186/s12866-014-0332-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Correa-Galeote D, Bedmar EJ, Arone GJ. Maize endophytic bacterial diversity as affected by soil cultivation history. Front. Microbiol. 2018;9:484. doi: 10.3389/fmicb.2018.00484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abdelrazek S, et al. Crop management system and carrot genotype affect endophyte composition and Alternaria dauci suppression. PLoS ONE. 2020;15(6):e0233783. doi: 10.1371/journal.pone.0233783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Horsfall JG. An improved grading system for measuring plant diseases. Phytopathology. 1945;35:655. [Google Scholar]
- 50.Brown, J.R., Recommended chemical soil test procedures for the North Central Region. 1998: Missouri Agricultural Experiment Station, University of Missouri-Columbia
- 51.Green VS, Stott DE, Diack M. Assay for fluorescein diacetate hydrolytic activity: Optimization for soil samples. Soil Biol. Biochem. 2006;38(4):693–701. [Google Scholar]
- 52.Weil RR, et al. Estimating active carbon for soil quality assessment: A simplified method for laboratory and field use. Am. J. Alternat. Agric. 2003;18(1):3–17. [Google Scholar]
- 53.Buyer JS, Sasser M. High throughput phospholipid fatty acid analysis of soils. Appl. Soil. Ecol. 2012;61:127–130. [Google Scholar]
- 54.Institute, S., JMP: Statistics and Graphics Guide. 2000: Sas Inst.
- 55.Surette MA, et al. Bacterial endophytes in processing carrots (Daucus carota L. var. sativus): Their localization, population density, biodiversity and their effects on plant growth. Plant Soil. 2003;253(2):381–390. [Google Scholar]
- 56.Corry, J.E., G.D. Curtis, and R.M. Baird, Handbook of culture media for food and water microbiology. 2011: Royal Society of Chemistry.
- 57.Reasoner DJ, Geldreich E. A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 1985;49(1):1–7. doi: 10.1128/aem.49.1.1-7.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993;2(2):113–118. doi: 10.1111/j.1365-294x.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 59.White TJ, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. 1990;18(1):315–322. [Google Scholar]
- 60.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7(5):335. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 62.Tikhonov M, Leach RW, Wingreen NS. Interpreting 16S metagenomic data without clustering to achieve sub-OTU resolution. ISME J. 2015;9(1):68–80. doi: 10.1038/ismej.2014.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Faith DP. Conservation evaluation and phylogenetic diversity. Biol. Cons. 1992;61(1):1–10. [Google Scholar]
- 64.Chao, A., Nonparametric estimation of the number of classes in a population. Scandinavian Journal of statistics, 265–270 (1984).
- 65.Faith DP, Baker AM. Phylogenetic diversity (PD) and biodiversity conservation: Some bioinformatics challenges. Evol. Bioinform. 2006;2:117693430600200007. [PMC free article] [PubMed] [Google Scholar]
- 66.Vázquez-Baeza Y, et al. EMPeror: A tool for visualizing high-throughput microbial community data. Gigascience. 2013;2(1):16. doi: 10.1186/2047-217X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26(1):32–46. [Google Scholar]
- 68.Roberts, D.W. and M.D.W. Roberts, Package ‘labdsv’. Ordination and Multivariate, 2016.
- 69.Hill, M., R. Bunce, and M. Shaw, Indicator species analysis, a divisive polythetic method of classification, and its application to a survey of native pinewoods in Scotland. The Journal of Ecology, 597–613 (1975).
- 70.Du Toit L, et al. First report of bacterial blight of carrot in Indiana caused by Xanthomonas hortorum pv. carotae. Plant Dis. 2014;98(5):685–685. doi: 10.1094/PDIS-10-13-1083-PDN. [DOI] [PubMed] [Google Scholar]
- 71.Arnold AE, Herre EA. Canopy cover and leaf age affect colonization by tropical fungal endophytes: Ecological pattern and process in Theobroma cacao (Malvaceae) Mycologia. 2003;95(3):388–398. [PubMed] [Google Scholar]
- 72.Gazis R, Chaverri P. Diversity of fungal endophytes in leaves and stems of wild rubber trees (Hevea brasiliensis) in Peru. Fungal Ecol. 2010;3(3):240–254. [Google Scholar]
- 73.Rivera-Orduña FN, et al. Diversity of endophytic fungi of Taxus globosa (Mexican yew) Fungal Divers. 2011;47(1):65–74. [Google Scholar]
- 74.Vieira ML, et al. Diversity and antimicrobial activities of the fungal endophyte community associated with the traditional Brazilian medicinal plant Solanum cernuum Vell. (Solanaceae) Can. J. Microbiol. 2011;58(1):54–66. doi: 10.1139/w11-105. [DOI] [PubMed] [Google Scholar]
- 75.Singh DK, et al. Diversity of endophytic mycobiota of tropical tree Tectona grandis Linn. f.: Spatiotemporal and tissue type effects. Sci. Rep. 2017;7:2. doi: 10.1038/s41598-017-03933-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Arnold AE, et al. Fungal endophytes limit pathogen damage in a tropical tree. Proc. Natl. Acad. Sci. 2003;100(26):15649–15654. doi: 10.1073/pnas.2533483100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pel MJ, Pieterse CM. Microbial recognition and evasion of host immunity. J. Exp. Bot. 2013;64(5):1237–1248. doi: 10.1093/jxb/ers262. [DOI] [PubMed] [Google Scholar]
- 78.Arnold A, et al. Hyperdiverse fungal endophytes and endolichenic fungi elucidate the evolution of major ecological modes in the Ascomycota. Syst. Biol. 2009;58:283–297. [Google Scholar]
- 79.Li H-Y, et al. Endophytes and their role in phytoremediation. Fungal Divers. 2012;54(1):11–18. [Google Scholar]
- 80.Wang F, et al. Arbuscular mycorrhizae alleviate negative effects of zinc oxide nanoparticle and zinc accumulation in maize plants–a soil microcosm experiment. Chemosphere. 2016;147:88–97. doi: 10.1016/j.chemosphere.2015.12.076. [DOI] [PubMed] [Google Scholar]
- 81.Nilsson RH, et al. The ITS region as a target for characterization of fungal communities using emerging sequencing technologies. FEMS Microbiol. Lett. 2009;296(1):97–101. doi: 10.1111/j.1574-6968.2009.01618.x. [DOI] [PubMed] [Google Scholar]
- 82.Motooka D, et al. Fungal ITS1 deep-sequencing strategies to reconstruct the composition of a 26-species community and evaluation of the gut mycobiota of healthy Japanese individuals. Front. Microbiol. 2017;8:238. doi: 10.3389/fmicb.2017.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dumas-Gaudot E, et al. A technical trick for studying proteomics in parallel to transcriptomics in symbiotic root-fungus interactions. Proteomics. 2004;4(2):451–453. doi: 10.1002/pmic.200300627. [DOI] [PubMed] [Google Scholar]
- 84.Kõljalg U, et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013;22(21):5271–5277. doi: 10.1111/mec.12481. [DOI] [PubMed] [Google Scholar]
- 85.Lemanceau P, et al. Let the core microbiota be functional. Trends Plant Sci. 2017;22(7):583–595. doi: 10.1016/j.tplants.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 86.Shade A, Handelsman J. Beyond the Venn diagram: The hunt for a core microbiome. Environ. Microbiol. 2012;14(1):4–12. doi: 10.1111/j.1462-2920.2011.02585.x. [DOI] [PubMed] [Google Scholar]
- 87.Pancher, M., et al., Fungal endophytic communities in grapevines (Vitis vinifera L.) respond to crop management. Applied and environmental microbiology, 2012: p. AEM. 07655–11. [DOI] [PMC free article] [PubMed]
- 88.Xia Y, et al. Characterization of culturable bacterial endophytes and their capacity to promote plant growth from plants grown using organic or conventional practices. Front. Plant Sci. 2015;6:490. doi: 10.3389/fpls.2015.00490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Reeve J, et al. Advances in Agronomy. Amserdam: Elsevier; 2016. Organic farming, soil health, and food quality: considering possible links; pp. 319–367. [Google Scholar]
- 90.Hoagland L, et al. Orchard floor management effects on nitrogen fertility and soil biological activity in a newly established organic apple orchard. Biol. Fertil. Soils. 2008;45(1):11. [Google Scholar]
- 91.Rudisill MA, et al. Sustaining soil quality in intensively managed high tunnel vegetable production systems: A role for green manures and chicken litter. HortScience. 2015;50(3):461–468. [Google Scholar]
- 92.Seghers D, et al. Impact of agricultural practices on the Zea mays L. endophytic community. Appl. Environ. Microbiol. 2004;70(3):1475–1482. doi: 10.1128/AEM.70.3.1475-1482.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen S, Reese CD. Parasitism of the nematode Heterodera glycines by the fungus Hirsutella rhossiliensis as influenced by crop sequence. J. Nematol. 1999;31(4):437. [PMC free article] [PubMed] [Google Scholar]
- 94.D'Amico M, Frisullo S, Cirulli M. Endophytic fungi occurring in fennel, lettuce, chicory, and celery—commercial crops in southern Italy. Mycol. Res. 2008;112(1):100–107. doi: 10.1016/j.mycres.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 95.González-Teuber M, Vilo C, Bascuñán-Godoy L. Molecular characterization of endophytic fungi associated with the roots of Chenopodium quinoa inhabiting the Atacama Desert, Chile. Genom. Data. 2017;11:109–112. doi: 10.1016/j.gdata.2016.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Riches, M.R.M.J.V.K., Muck Vegetable Cultivar Trial& Research Report2016. 2016.
- 97.Liu B, et al. Effect of organic, sustainable, and conventional management strategies in grower fields on soil physical, chemical, and biological factors and the incidence of Southern blight. Appl. Soil. Ecol. 2007;37(3):202–214. [Google Scholar]
- 98.van Bruggen AH, et al. Soil health indicators and Fusarium wilt suppression in organically and conventionally managed greenhouse soils. Appl. Soil. Ecol. 2015;86:192–201. [Google Scholar]
- 99.Cai X, et al. Long-term organic farming manipulated rhizospheric microbiome and Bacillus antagonism against Pepper blight (Phytophthora capsici) Front. Microbiol. 2019;10:342. doi: 10.3389/fmicb.2019.00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu H, et al. Dark septate endophytes colonizing the roots of ‘non-mycorrhizal’plants in a mine tailing pond and in a relatively undisturbed environment, Southwest China. J. Plant Interact. 2017;12(1):264–271. [Google Scholar]
- 101.Bonito G, et al. Plant host and soil origin influence fungal and bacterial assemblages in the roots of woody plants. Mol. Ecol. 2014;23(13):3356–3370. doi: 10.1111/mec.12821. [DOI] [PubMed] [Google Scholar]
- 102.Chen YH, Gols R, Benrey B. Crop domestication and its impact on naturally selected trophic interactions. Annu. Rev. Entomol. 2015;60:35–58. doi: 10.1146/annurev-ento-010814-020601. [DOI] [PubMed] [Google Scholar]
- 103.Williamson, V.M., P.A. Roberts, and R. Perry, 13. Mechanisms and genetics of resistance. Root-knot nematodes, 2009. 301.
- 104.Davis RM. Diseases of Fruits and Vegetables. Berlin: Springer; 2004. Carrot diseases and their management; pp. 397–439. [Google Scholar]
- 105.Farrokhi-Nejad R, Cromey MG, Moosawi-Jorf SA. Determination of the anastomosis grouping and virulence of Rhizoctonia spp. associated with potato tubers grown in Lincoln, New Zealand. Pak. J. Biol. Sci. 2007;10(21):3786–3793. doi: 10.3923/pjbs.2007.3786.3793. [DOI] [PubMed] [Google Scholar]
- 106.Durán-López M, et al. The micorryzal fungi Ceratobasidium sp. and Sebacina vermifera promote seed germination and seedling development of the terrestrial orchid Epidendrum secundum Jacq. South Afr. J. Bot. 2019;125:54–61. [Google Scholar]
- 107.Mosquera-Espinosa AT, et al. The double life of Ceratobasidium: orchid mycorrhizal fungi and their potential for biocontrol of Rhizoctonia solani sheath blight of rice. Mycologia. 2013;105(1):141–150. doi: 10.3852/12-079. [DOI] [PubMed] [Google Scholar]
- 108.Vanhove W, Vanhoudt N, Van Damme P. Biocontrol of vascular streak dieback (Ceratobasidium theobromae) on cacao (Theobroma cacao) through induced systemic resistance and direct antagonism. Biocontrol Sci. Tech. 2016;26(4):492–503. [Google Scholar]
- 109.Taufik, M., et al. Evaluating the ability of endophyte fungus to tontrol VSD diseases in cocoa seeding. in IOP Conference Series: Earth and Environmental Science. 2019. IOP Publishing.
- 110.du Toit L, Derie M. First report of Cladosporium leaf spot of spinach caused by Cladosporium variabile in the winter spinach production region of California and Arizona. Plant Dis. 2012;96(7):1071–1071. doi: 10.1094/PDIS-03-12-0279-PDN. [DOI] [PubMed] [Google Scholar]
- 111.Hamayun M, et al. Gibberellin production by pure cultures of a new strain of Aspergillus fumigatus. World J. Microbiol. Biotechnol. 2009;25(10):1785–1792. [Google Scholar]
- 112.Nesha R, Siddiqui ZA. Effects of Paecilomyces lilacinus and Aspergillus niger alone and in combination on the growth, chlorophyll contents and soft rot disease complex of carrot. Sci. Hortic. 2017;218:258–264. [Google Scholar]
- 113.Wang F, et al. Antimicrobial potentials of endophytic fungi residing in Quercus variabilis and brefeldin A obtained from Cladosporium sp. World J. Microbiol. Biotechnol. 2007;23(1):79–83. [Google Scholar]
- 114.Li X-J, et al. Metabolites from Aspergillus fumigatus, an endophytic fungus associated with Melia azedarach, and their antifungal, antifeedant, and toxic activities. J. Agric. Food Chem. 2012;60(13):3424–3431. doi: 10.1021/jf300146n. [DOI] [PubMed] [Google Scholar]
- 115.de Vries RP, de Lange ES, Stalpers JA. Control and possible applications of a novel carrot-spoilage basidiomycete, Fibulorhizoctoniaápsychrophila. Antonie Van Leeuwenhoek. 2008;93(4):407–413. doi: 10.1007/s10482-007-9218-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Iturralde Martinez JF, et al. Multiplex end-point PCR for the detection of three species of ophiosphaerella causing spring dead spot of bermudagrass. Plant Dis. 2019;103(8):2010–2014. doi: 10.1094/PDIS-10-18-1727-RE. [DOI] [PubMed] [Google Scholar]
- 117.Gao, L., et al., Three new species of Cyphellophora (Chaetothyriales) associated with sooty blotch and flyspeck. PLoS One, 2015. 10(9). [DOI] [PMC free article] [PubMed]
- 118.Spagnoletti F, et al. Dark septate endophytes present different potential to solubilize calcium, iron and aluminum phosphates. Appl. Soil. Ecol. 2017;111:25–32. [Google Scholar]
- 119.Vayssier-Taussat M, et al. Shifting the paradigm from pathogens to pathobiome: New concepts in the light of meta-omics. Front. Cell. Infect. Microbiol. 2014;4:29. doi: 10.3389/fcimb.2014.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lee K, Pan JJ, May G. Endophytic Fusarium verticillioides reduces disease severity caused by Ustilago maydis on maize. FEMS Microbiol. Lett. 2009;299(1):31–37. doi: 10.1111/j.1574-6968.2009.01719.x. [DOI] [PubMed] [Google Scholar]
- 121.Aimé S, et al. The endophytic strain Fusarium oxysporum Fo47: a good candidate for priming the defense responses in tomato roots. Mol. Plant Microbe Interact. 2013;26(8):918–926. doi: 10.1094/MPMI-12-12-0290-R. [DOI] [PubMed] [Google Scholar]
- 122.CaféFilho A, Reifschneider F, Tateishi NT. Pathogenicity of Colletotrichum gloeosporioides to carrot. Int. J. Pest Manag. 1986;32(4):274–276. [Google Scholar]
- 123.Redman RS, et al. Biochemical analysis of plant protection afforded by a nonpathogenic endophytic mutant of Colletotrichum magna. Plant Physiol. 1999;119(2):795–804. doi: 10.1104/pp.119.2.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lu H, et al. New bioactive metabolites produced by Colletotrichum sp., an endophytic fungus in Artemisia annua. Plant Sci. 2000;151(1):67–73. [Google Scholar]
- 125.Hiruma K, et al. Root endophyte Colletotrichum tofieldiae confers plant fitness benefits that are phosphate status dependent. Cell. 2016;165(2):464–474. doi: 10.1016/j.cell.2016.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.