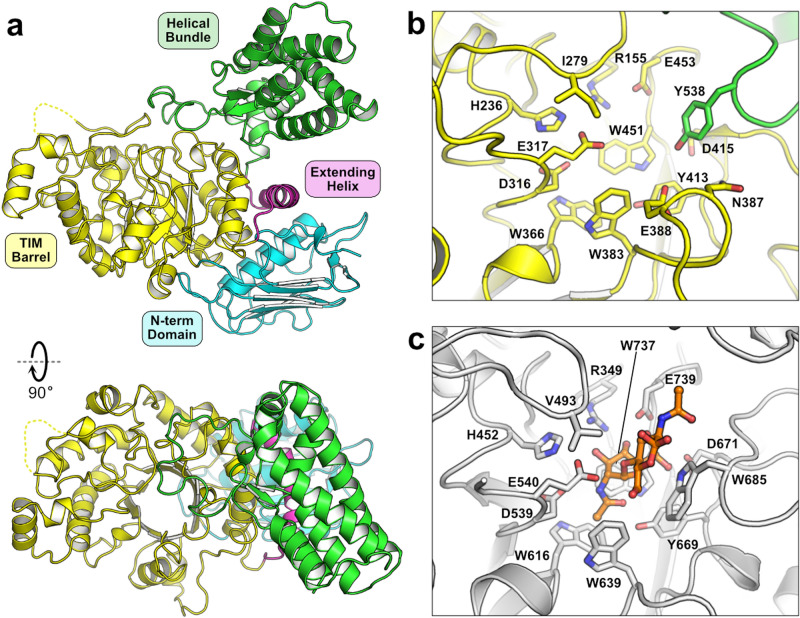
Figure 1.
Structure of FjGH20. The overall structure of the enzyme is shown with the individual domains annotated by color (a). The active site architectures of FjGH20 (b) and SmGH20 (c; PDB accession 1qbb) are shown with key residues lining the substrate-binding pocket. The structure of SmGH2020 was determined in complex with chitobiose (orange carbons). A notable difference between the two enzymes is the presence of a tyrosine (Tyr538) from a loop of the helical bundle domain in FjGH20 filling the void left by the absence of a conserved tryptophan (Trp685 in SmGH20), which is involved in substrate binding by ring stacking with a GlcNAc residue in chitobiose.