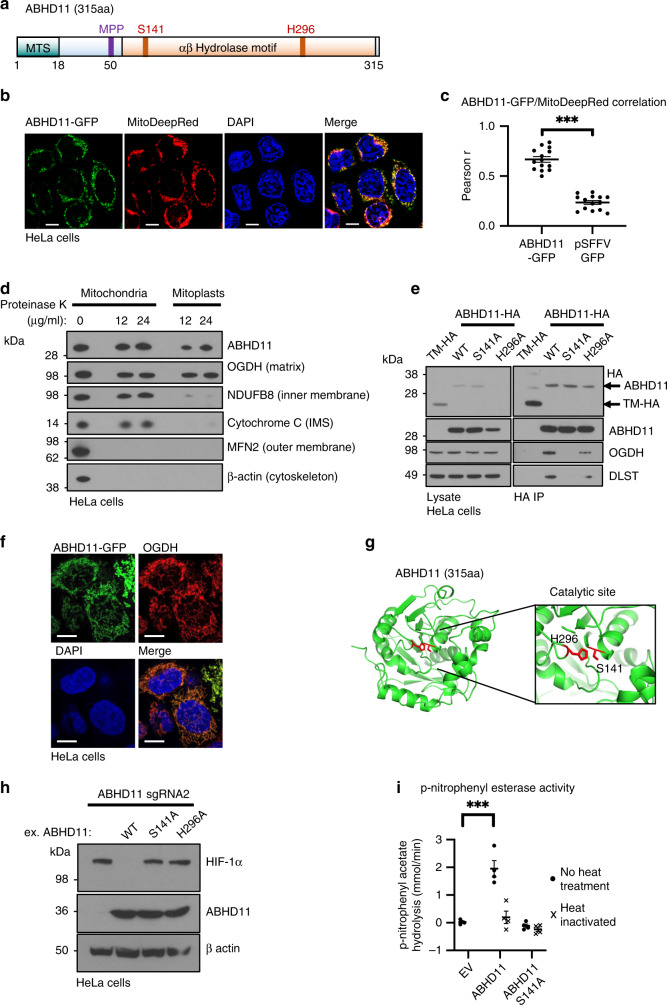
Fig. 3. ABHD11 is a serine hydrolase that associates with the OGDHc.
a Schematic of ABHD11 with the putative mitochondrial targeting sequence (MTS), mitochondrial processing peptidase (MPP) cleavage site and catalytic residues indicated (Serine 141, and Histidine 296). Modelled from UniProtKB—Q8NFV4, and MitoFates prediction tool55. b Confocal micrograph of HeLa cells lentivirally transduced with ABHD11-GFP. Mitochondria were visualised with MitoTracker Deep Red FM (MitoDeepRed); Scale = 10 μm, representative example from two biologically independent experiments and 14 images (c) Pearson correlation coefficient (Pearson r) comparing colocalisation of GFP and MitoTracker in HeLa cells expressing ABHD11-GFP (n = 14). HeLa cells expressing GFP under an SFFV promoter without a localisation signal served as a cytosolic control (pSFFV-GFP; n = 14) ***p = 7.0 × 10−6, two-tailed Mann–Whitney U test. d Mitochondrial protease protection assay. Mitochondria were extracted using the Qproteome Mitochondria Isolation Kit (Qiagen). Proteinase K was added to the final concentrations indicated, and incubation at 37 °C for 30 min. e Immunoprecipitation of ABHD11-HA with endogenous OGDHc components. ABHD11-HA or the inactive mutants (S141A and H296A) were transduced into HeLa cells, lysed and immunoprecipitated using the HA tag. TMEM199, a membrane bound protein tagged with HA (TM-HA) was used as a control. f Colocalisation of ABHD11 with the mitochondrial matrix protein, OGDH. HeLa cells expressing ABHD11-GFP were fixed in paraformaldehyde. ABHD11 and OGDH subcellular localisation was visualised by immunofluorescence confocal microscopy. Scale = 10 μm, representative image of five technical repeats. g In silico modelling of ABHD11 with putative catalytic site and key residues S141 and H296 (Phyre2 structural prediction against a template of murine epoxide hydrolase, PDB: 1cr6, and visualised using PyMOL 2.3). h Reconstitution of mixed KO population of ABHD11 with exogenous ABHD11, or enzymatic inactive mutants. i p-nitrophenyl esterase activity of purified ABHD11-FLAG. Purified wildtype or S141A ABHD11-FLAG were incubated with p-nitrophenyl acetate and hydrolysis measured by rate of increase in absorbance at 405 nm (37 °C for 40 min). An empty FLAG vector (EV), that had undergone affinity purification, was used as a control. ABHD11 enzymatic activity was also measured following heat inactivation of the protein (90 °C for 5 min). n = 4, Mean ± SEM, ***p = 0.0006, two-tailed t test.