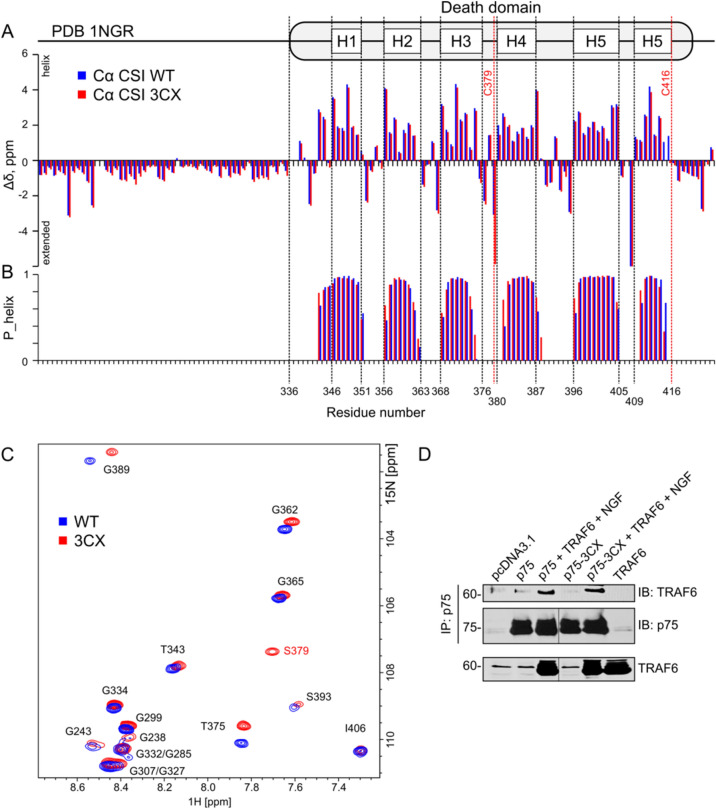
Figure 4.
Comparison of structure and activity of wt and 3CX mutant forms of rP75NTR. (A) Secondary chemical shifts of Cα nuclei of rP75-ΔECD (blue bars) and rP75-ΔECD-3CX (red bars). The secondary structure of the death domain, according to PDB ID 1NGR is shown on top. (B) The propensity of ɑ-helical conformation, according to the TALOS-N prediction, based on NMR chemical shifts of rP75-ΔECD (blue bars) and rP75-ΔECD-3CX (red bars). (C) Overlay of 1H,15N-TROSY-HSQC spectra (glycine regions) of rP75-ΔECD (blue) and rP75-ΔECD-3CX (red). The assignment is provided in black and red (for the mutated S379 residue). (D) Binding of TRAF6 to wild type and mutant rP75-3CX in transfected HEK293 cells. Full-length blots are presented in Supplementary Fig S8. The figure was prepared using the program Inkscape 0.92 (https://inkscape.org/).