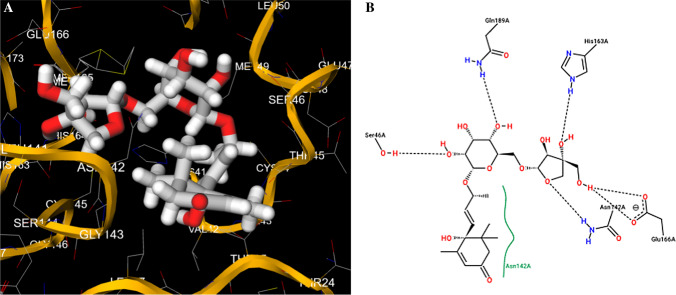
Fig. 4.
Putative best docking pose (a) and 2D protein–ligand interaction diagram (b) at the active site of main protease Mpro for lauruside 5. Proximity of functional groups of the laurel glycoside to Ser46, Asn142, Glu166 protein residues is observed in the pose view (a), while main interactions, including H-bonds with amino acids Ser46, Gln189, His163, Asn142, Glu166, are evidenced in the diagram (b). Note the highly stabilized nature of the predicted lauruside-protease complex due to the multiple H-bonding possibilities given by the interactions of the compound hydroxyl moieties with the protein amino acids