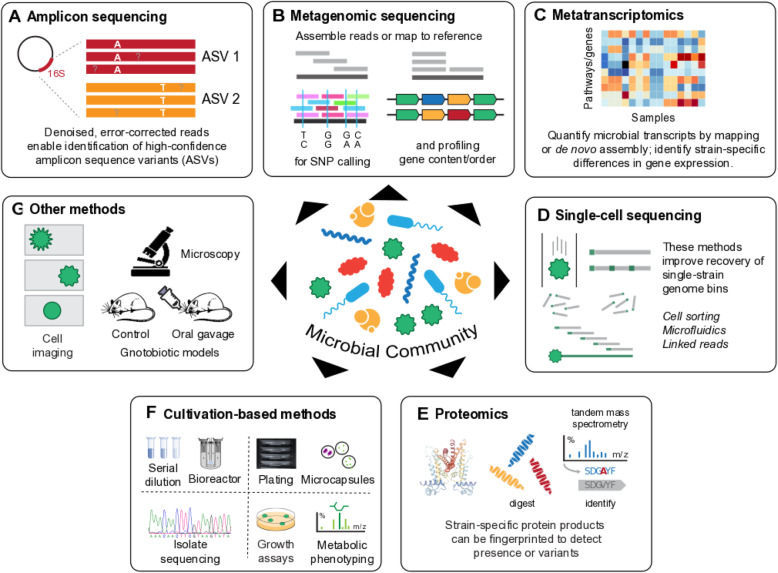
Fig. 1.
Strain identification approaches for microbial communities. This review summarizes a variety of high-throughput, often (but not always) culture-independent methods for strain identification within microbial communities. a Amplicon sequencing (e.g., 16S rRNA gene regions) can now be processed to near-strain-level fidelity, resulting in unique markers such as amplicon sequence variants (ASVs). b Shotgun metagenomic sequencing, either via assembly or using reference-based approaches, can identify strains broadly based on their single-nucleotide variants (SNVs) or structural variants (gene gain and loss events). c Whole-community transcriptomes can amplify the effects of gene gains or losses, or the effects of small variants that result in differential expression. d Single-cell methods can isolate individual microbial genomics directly from within communities, either via cell sorting and amplification, or through synthetic long-read/linked-read techniques. e High-throughput “culturomics” can be combined with rapid turnaround approaches such as peptide fingerprinting to strain-type isolates or microcolonies. f Relatedly, any combination of traditional isolation and high-throughput cultivation—batch, serial, or continuous—can be combined with growth, phenotypic, or molecular readouts for strain identification. g Finally, a variety of other approaches can be used with communities, ranging from flow- or high-content microscopic imaging to systems such as gnotobiotic animal model physiology and phenotyping