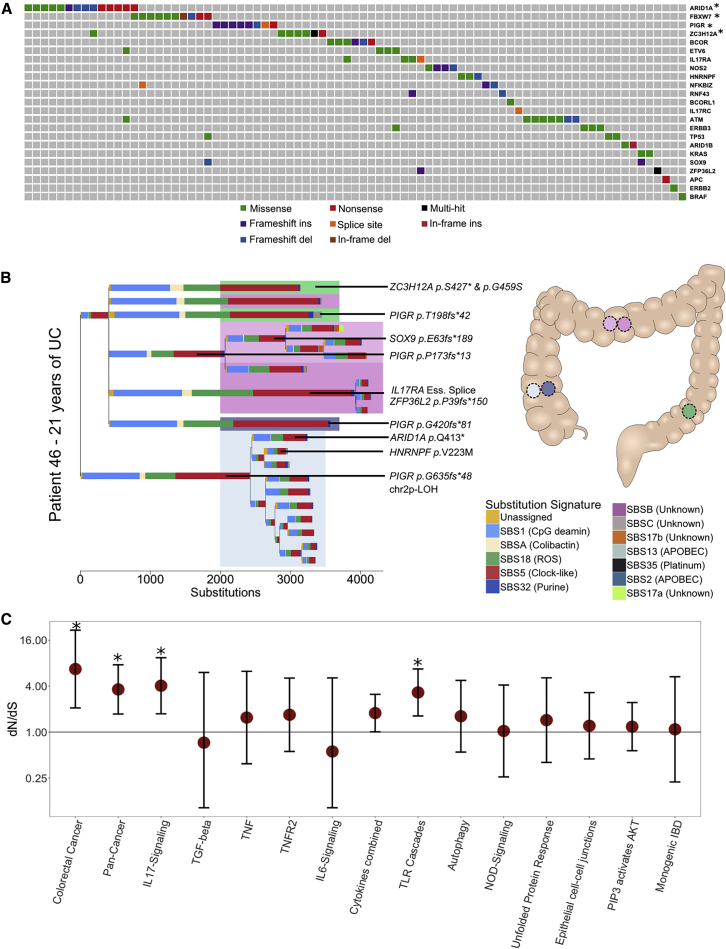
Figure 6.
Driver Mutations and Positive Selection in IBD
(A) An oncoplot showing the distribution of potential driver mutations mapped to branches of phylogenetic trees. Each column represents a branch of a phylogenetic tree and a mutation may be found in multiple crypts if the branch precedes a clonal expansion. Branches without potential drivers are not shown for simplicity. ∗Genes showing significant enrichment of non-synonymous mutations after Benjamini-Hochberg correction for multiple testing (q < 0.05).
(B) A phylogenetic tree of the crypts dissected from a 38-year-old male suffering from ulcerative colitis for 21 years. Crypts are dissected from five biopsies from three previously inflamed sites of the colon. Crypts carrying distinct PIGR truncating mutations are found in four of the biopsies and in all three colonic sites.
(C) Pathway-level dN/dS ratios for truncating mutations in known cancer genes and cellular pathways important in IBD pathogenesis. Error bars represent 95% confidence intervals. *q < 0.05 after Benjamini-Hochberg correction for multiple testing.