Figure 4.
Mathematical Model of RNAPI Transcription
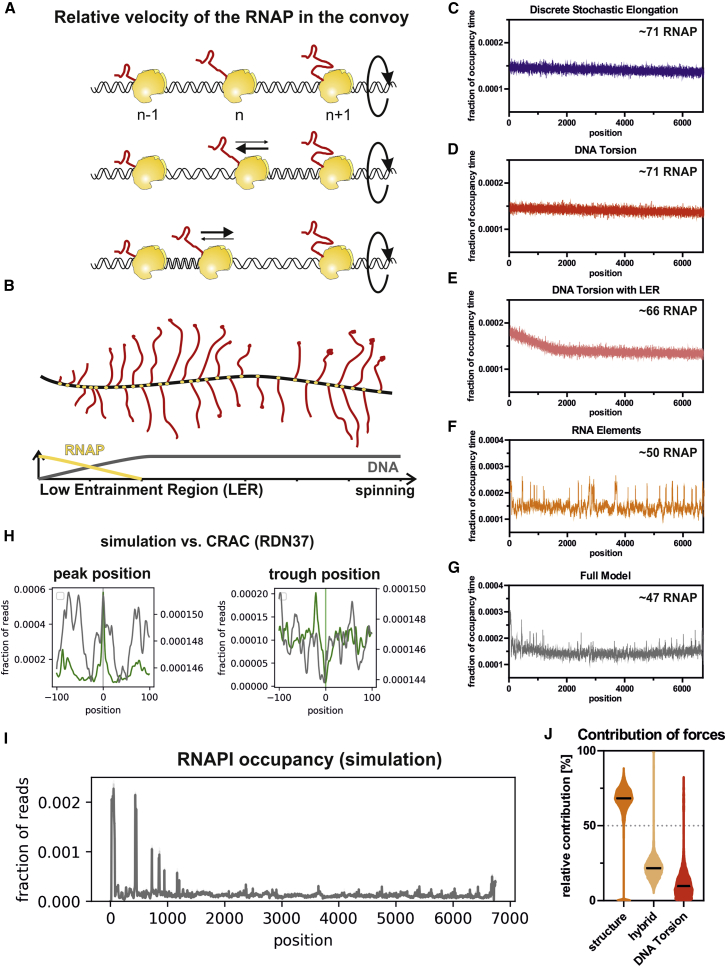
(A) Schematic of polymerase convoys in which a group of RNAP complexes moves along the rDNA transcription unit while DNA screws through the polymerases. In this model, the distance between initially loaded RNAP molecules is retained by the DNA helix, which behaves as an elastic rod and generates force when over- or under-wound.
(B) Schematic representation of the low entrainment region (LER). RNAPI molecules are initially able to rotate around the DNA, allowing changes in their relative positions without generating torsion. Grey line, degree of torsional entrainment; yellow line, ability of RNAPI to reduce torsion by rotation around the rDNA.
(C) Modeled RNAPI occupancy along the transcription unit using a model of stochastic initiation and discrete, stochastic elongation. The average number of RNAP molecules per transcription unit is indicated. The average RNAPI occupancy for 64 simulations is presented. Each simulation was run for 2,000 sec and 200 time points were collected (C–G).
(D) Modeled RNAPI occupancy for the DNA torsion model.
(E) Modeled RNAPI occupancy for the DNA torsion model, including a LER of 2,000 nt. DNA torsion is engaged linearly between positions 0 and 2,000.
(F) RNAPI occupancy for the model with RNA elements. Translocation is modified positively by structure in the nascent RNA extruded from the polymerase and negatively by the stability of the RNA:DNA hybrid within the transcription bubble.
(G) Full model including DNA torsion, the LER, and RNA elements. This recapitulates experimental data to the greatest extent. Areas that do not correlate with experimental data are predicted to be affected by trans-acting factors binding co-transcriptionally (Discussion).
(H) RNAPI CRAC peak and trough metaplots together with simulated data. Shown is a full model (gray) relative to CRAC data (green).
(I) Modeled RNAPI occupancy plot generated using the full model. Four datasets of 256 simulations were used to generate profiles. In silico “reads” were sampled from interpolated profiles, generated as in G, and processed in the same way as RNAPI CRAC data.
(J) Violin plot of factor contributions at each elongation step within the full model.
See also Figure S4.